DNA damage repair markers
Unrepaired DNA damage leads to mutagenesis or cell death.
DNA mutations are a known factor in the tumorigenesis of many cancer types including colon and skin cancer, leukemia, lymphoma, and breast or ovarian cancer.
The accumulation of DNA damage in myocytes or neurons can result in degenerative diseases (1). The human brain consumes 20% of the body’s entire oxygen intake to generate sufficient energy to maintain its proper functioning. Since ROS (Reactive Oxygen Species) are a natural side-product of the pathways of this energy production, they can be easily accumulated in the brain and induce DNA damage.
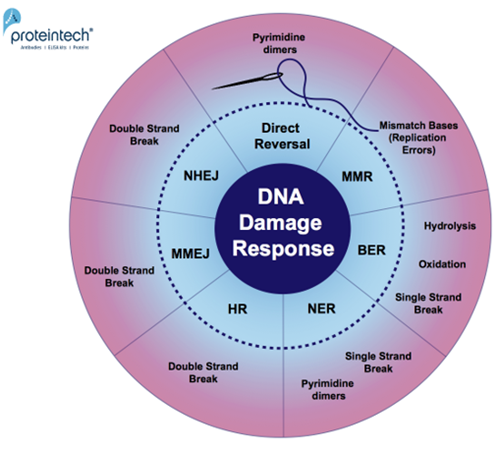
Every day, cells are exposed to a broad range of DNA-damaging factors (2). As a safeguard for genome stability, they have developed many repair pathways, especially for damage from oxidation, hydrolysis, dimerization of DNA-bases, and single or double strand DNA breaks (Figure 1).
|
|
| Figure 1. Overview of DNA damage-specific repair pathways. Response mechanisms: Direct reversal, MMR (MisMatch Repair), BER (Base Excision Repair), NER (Nucleotide Excision Repair), HR (Homologous Recombination), MMEJ (Microhomology-Mediated End Joining), and NHEJ (Non-Homologous End Joining). |
|
|
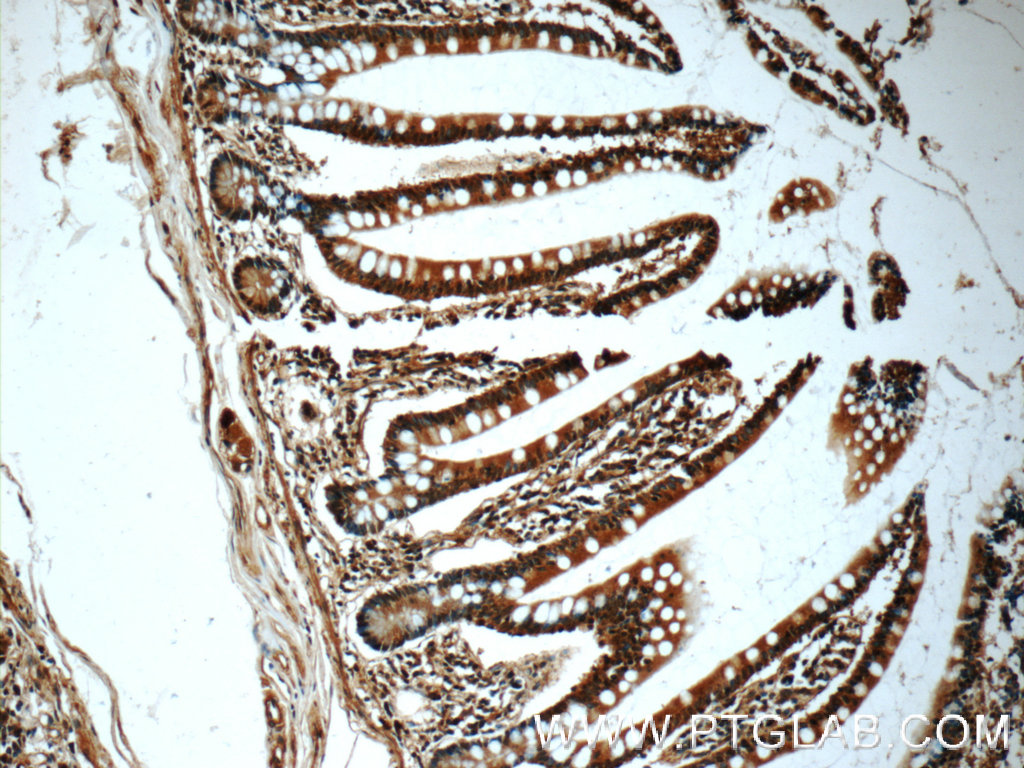
| Figure 2. Immunohistochemical analysis of paraffin-embedded human small intestine tissue slide using 60277-1-Ig ( CHK1 Antibody) at dilution of 1:50 (under 10x lens). |
Climate change and increased environmental pollution are extending DNA-damaging factors. It is therefore important to understand the DNA damage repair mechanisms. Tomas Lindahl, Paul Modrich, and Aziz Sancarwere pioneers in discovering the molecular basis of DNA damage repair pathways (2015, with the Nobel Prize in Chemistry).
 |
|
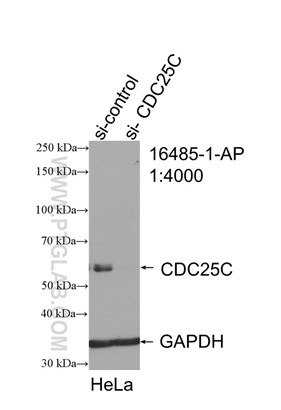
Figure 3. WB result of CDC25C antibody (16485-1-AP; 1:4000; incubated at room temperature for 1.5 hours) with sh-Control and sh-CDC25C transfected HeLa cells. |
 |
|
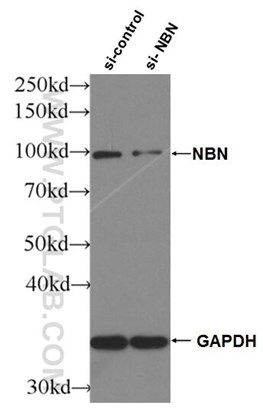
Figure 4. WB result of NBS1 antibody (55025-1-AP; 1:2000; incubated at room temperature for 1.5 hours) with sh-Control and sh-NBS1 transfected HeLa cells |
DNA damage repair markers
|
DNA damage marker |
Type |
Cat. No. |
Damage type/ Repair mechanism |
|
ATM |
Rabbit |
DSB |
|
|
ATR |
Rabbit |
SSB |
|
|
ATRIP |
Rabbit |
SSB |
|
|
Cdc25A |
Rabbit |
SSB, DSB |
|
|
Cdc25B |
Rabbit |
SSB, DSB |
|
|
Cdc25C |
Rabbit, Mouse |
16485-1-AP (Figure 3) ; 66912-1-Ig |
SSB, DSB |
|
Chk1 |
Rabbit, Mouse |
25667-1-AP; 60277-1-Ig (Figure 2) |
SSB |
|
Chk2 |
Rabbit |
DSB |
|
|
DDB1 |
Rabbit, Mouse |
SSB/ NER |
|
|
DDB2 |
Rabbit |
SSB/ NER |
|
|
Fen1 |
Rabbit |
DSB/ MMEJ |
|
|
H2AX |
Rabbit |
DSB |
|
|
Ku70 |
Rabbit, Mouse |
DSB/ NHEJ |
|
|
Ku80 |
Rabbit, Mouse |
DSB/ NHEJ |
|
|
Mre11 |
Rabbit |
DSB/ HR |
|
|
Nbs1 |
Rabbit |
55025-1-AP (Figure 4) |
DSB/ HR |
|
PCNA |
Rabbit, Mouse |
SSB/ NER |
|
|
POLD1 |
Rabbit |
SSB |
|
|
POLD2 |
Rabbit |
SSB |
|
|
POLD3 |
Rabbit |
SSB |
|
|
POLD4 |
Rabbit |
SSB |
|
|
POLE3 |
Rabbit |
SSB |
|
|
POLK |
Rabbit |
SSB |
|
|
Rad1 |
Rabbit |
SSB |
|
|
Rad51 |
Rabbit, Mouse |
DSB/ HR |
|
|
RFC2 |
Rabbit |
SSB/ NER |
|
|
RFC3 |
Rabbit |
SSB/ NER |
|
|
RFC4 |
Rabbit |
SSB/ NER |
|
|
RFC5 |
Rabbit |
SSB/ NER |
|
|
RPA1 |
Rabbit |
SSB, DSB/ HR |
|
|
RPA2 |
Rabbit |
SSB, DSB/ HR |
|
|
RPA3 |
Rabbit |
SSB, DSB/ HR |
|
|
RPA4 |
Rabbit |
SSB, DSB/ HR |
Loading control antibodies
| GAPDH Antibody | 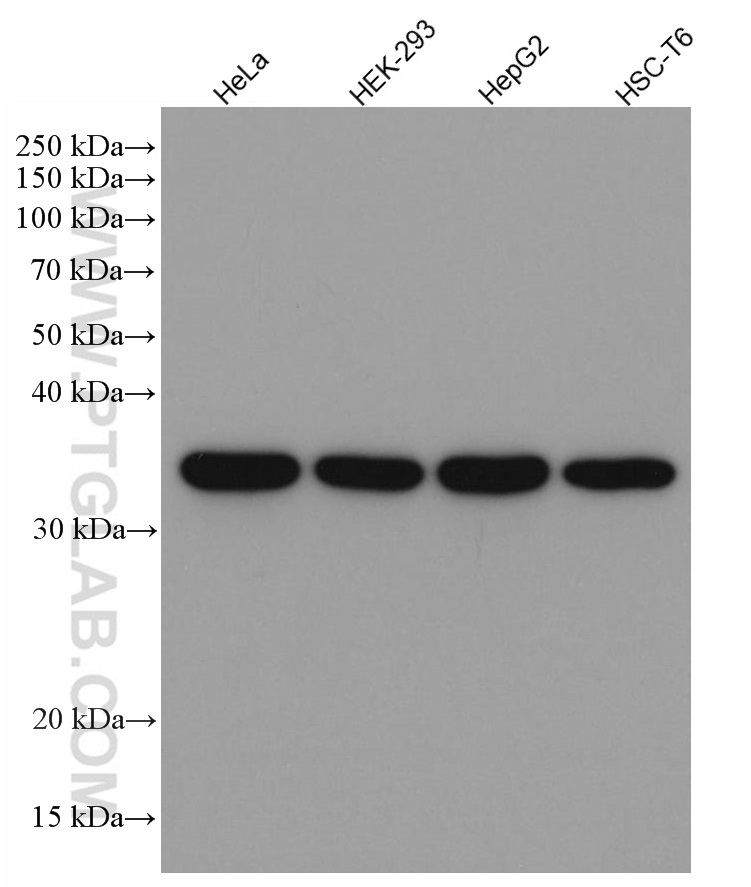 |
| Catalog no.: 60004-1-Ig | |
|
GAPDH is commonly used as a protein loading control in western blot due to its consistently high expression in most cell types. This enzyme participates in several cellular events such as glycolysis, DNA repair, and apoptosis. Proteintech monoclonal GAPDH antibodies are raised against a whole-protein antigen of human origin and have over 4,960 citations. |
| Beta Actin Antibody (KD/KO validated) | 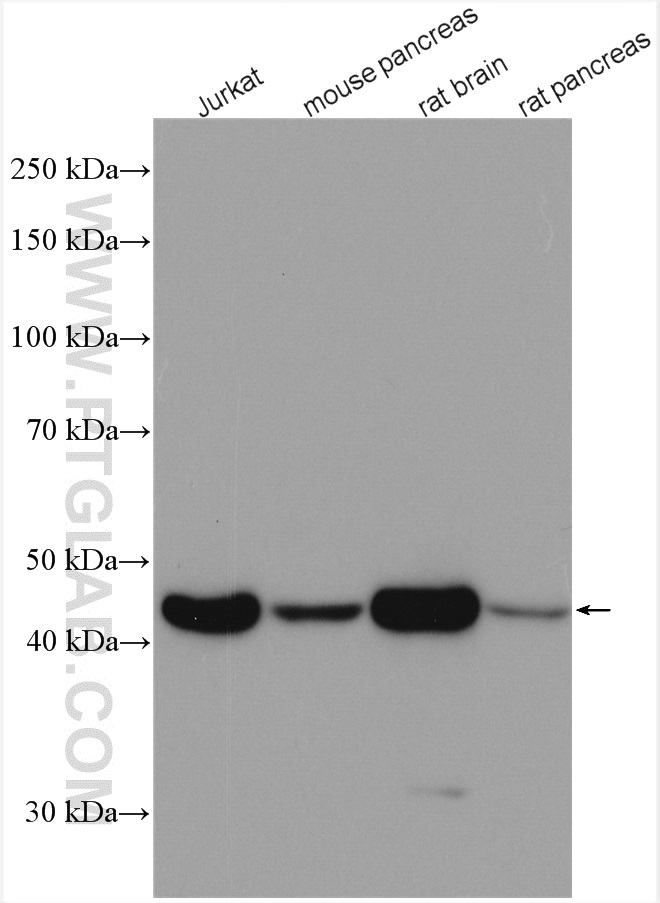 |
| Catalog no.: 66009-1-Ig | |
|
Beta-actin is usually used as a loading control due to its broad and consistent expression across all eukaryotic cell types and the fact that expression levels of this protein are not affected by most experimental treatments. 66009-1-Ig has been cited in over 2,460 publications and has wide species reactivity. |
Homologous Recombination (HR)
Table 1. List of available antibodies against DNA damage markers and the DNA damage pathway(s)
Double Strand Breaks (DSB)
Microhomology-Mediated End Joining (MMEJ)
Nucleotide Excision Repair (NER)
Non-Homologous End Joining (NHEJ)
Single Strand Breaks (SSB)
References
1. Stein and Toiber (2017) “DNA damage and neurodegeneration: the unusual suspect”.
2. Lindahl and Barnes (2000) “Repair of endogenous DNA damage“.