The Role of Autophagy in Cancer
Written by Dharaniya Sakthivel, PhD student at Baylor College of Medicine.
Autophagy, which means "self-eating," is a natural process wherein cells break down and recycle their components, including damaged organelles and misfolded proteins. Cells are often exposed to various physical, chemical, and biological stress stimuli that disrupt their homeostatic balance. Autophagy emerged as an intracellular adaptation mechanism to preserve cellular integrity. This is critical for cell survival in conditions of nutrient deprivation and infection. Autophagy also regulates various cellular processes, including cell growth, differentiation, and death.
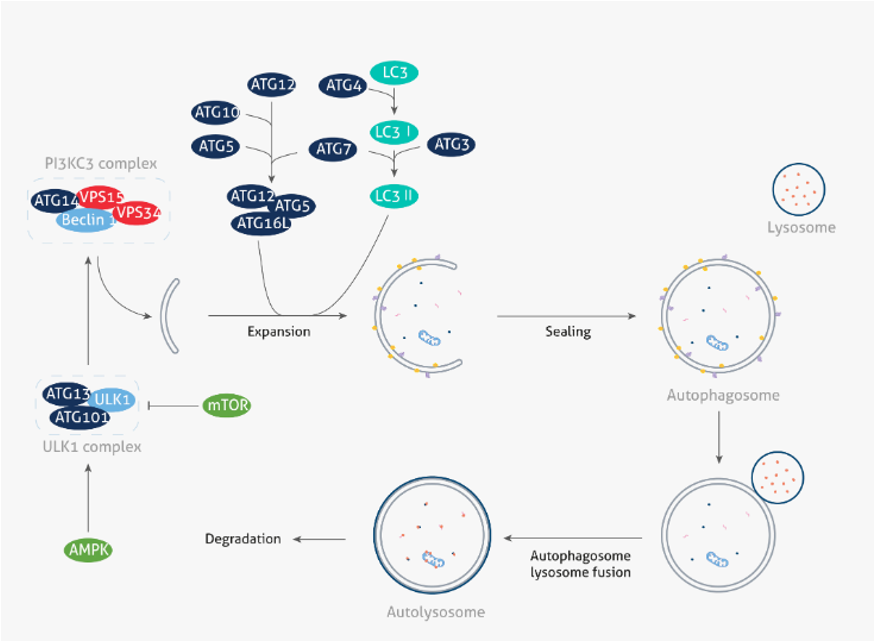
In normal cells, autophagy is strictly regulated by autophagy-related genes (ATG). The autophagy process typically occurs in three phases: 1) phagophore formation, 2) elongation and isolation, and 3) maturation. First, mammalian target of rapamycin (mTOR), the nutrient sensor interacts with the Unc-51-like kinase 1/2 (ULK1/2) complex. Under cellular stress, such as starvation, and AMP-activated protein kinase (AMPK), the energy sensor activates ULK for the generation of phagophores [1]. The next stage, elongation and isolation, is mediated by ATG5 - ATG12 complex and microtubule-associated protein 1A/1B-light chain 3 (LC3) pathways. Finally, in the maturation step, NSF attachment protein receptors (SNAREs) facilitate the formation of the autophagolysosome, which contains the engulfed particles for degradation and subsequent renewal. [2,3].
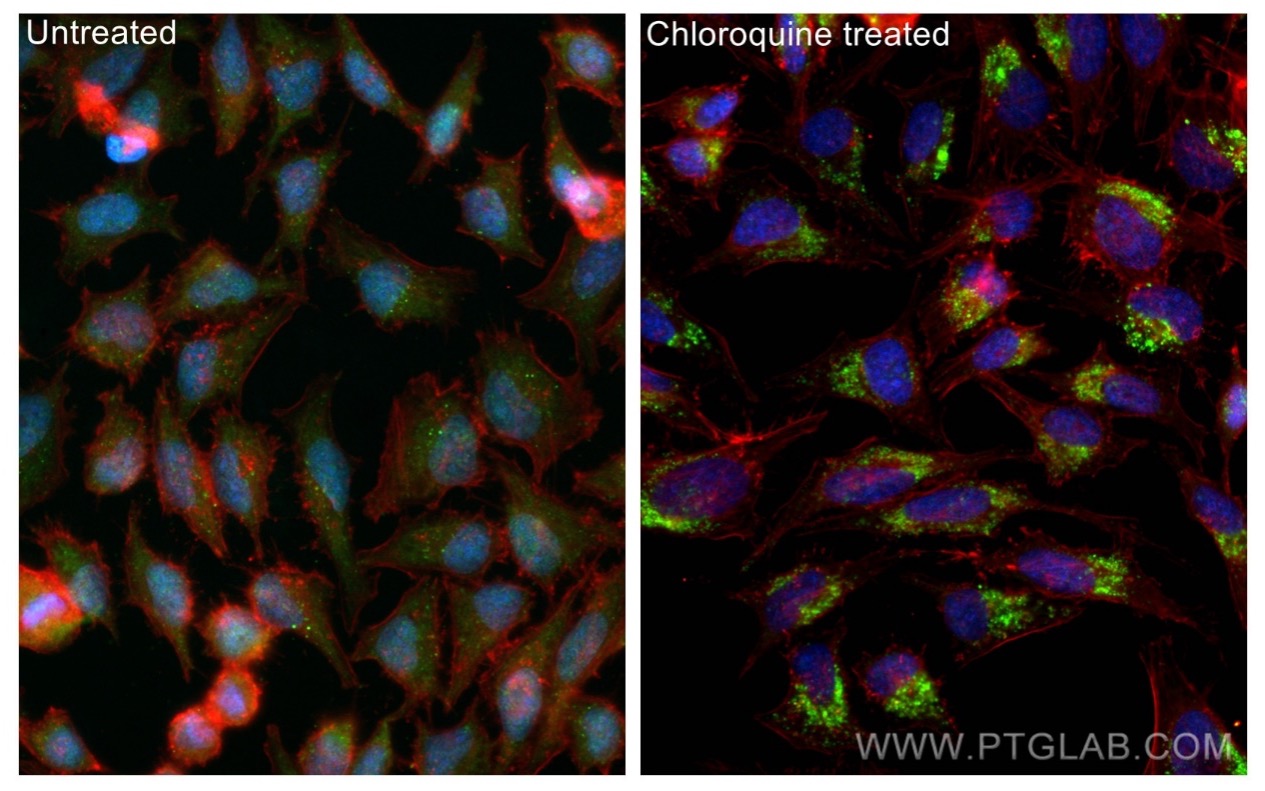
Immunofluorescence analysis of untreated and chloroquine treated Hela cells using recombinant LC3 antibody (81004-1-RR).
Proteintech’s Autophagy Antibody Kits provide cost-effective tools for studying key proteins involved in the autophagy pathway. While the Autophagy Essentials Antibody Kits are perfect for preliminary studies confirming the presence of autophagy in cells or tissue samples, the Autophagy Expanded Antibody Kits allow detailed analysis of the autophagy pathway.
Autophagy as a Tumor Suppressor
In cancer, autophagy is a double-edged sword and is highly dependent on the stage of the disease, the type of cancer, and the tumor microenvironment.
At basal levels, autophagy operates to suppress tumor growth due to its ability to remove dysfunctional cellular components and proteins. The autophagy-related gene BECN1, essential for phagophore formation, is depleted in breast, prostate, and ovarian cancers [4]. In cancer cell lines and mouse models, BECN1 loss reduces autophagy and promotes cell proliferation, suggesting that BECN1 is a potential tumor suppressor [5]. Moreover, proteins that associate with BECN1, such as UV radiation resistance-associated gene (UVRAG) and Bax-interacting factor-1 (Bif-1), function as a traditional tumor suppressor [6].
In mice, the deletion of autophagic core proteins, such as ATG5 and ATG7, results in the development of liver cancers from autophagy-deficient cells. ATG4 knockout in mice increases fibrosarcomas after exposure to chemical carcinogens. Such tumor growth is attributed to the accumulation of oxidative stress [7].
In addition, autophagy prevents tumor initiation through the clearing of damaged mitochondria, a significant source of reactive oxygen species (ROS) [8]. Increased ROS disrupts redox balance, activates pro-inflammatory signal cascades, enhances cell proliferation, and drives DNA damage and genomic instability contributing to tumor generation and progression. Taken together, autophagy is a crucial mechanism that serves to inhibit oncogenesis.
 |
 |
|
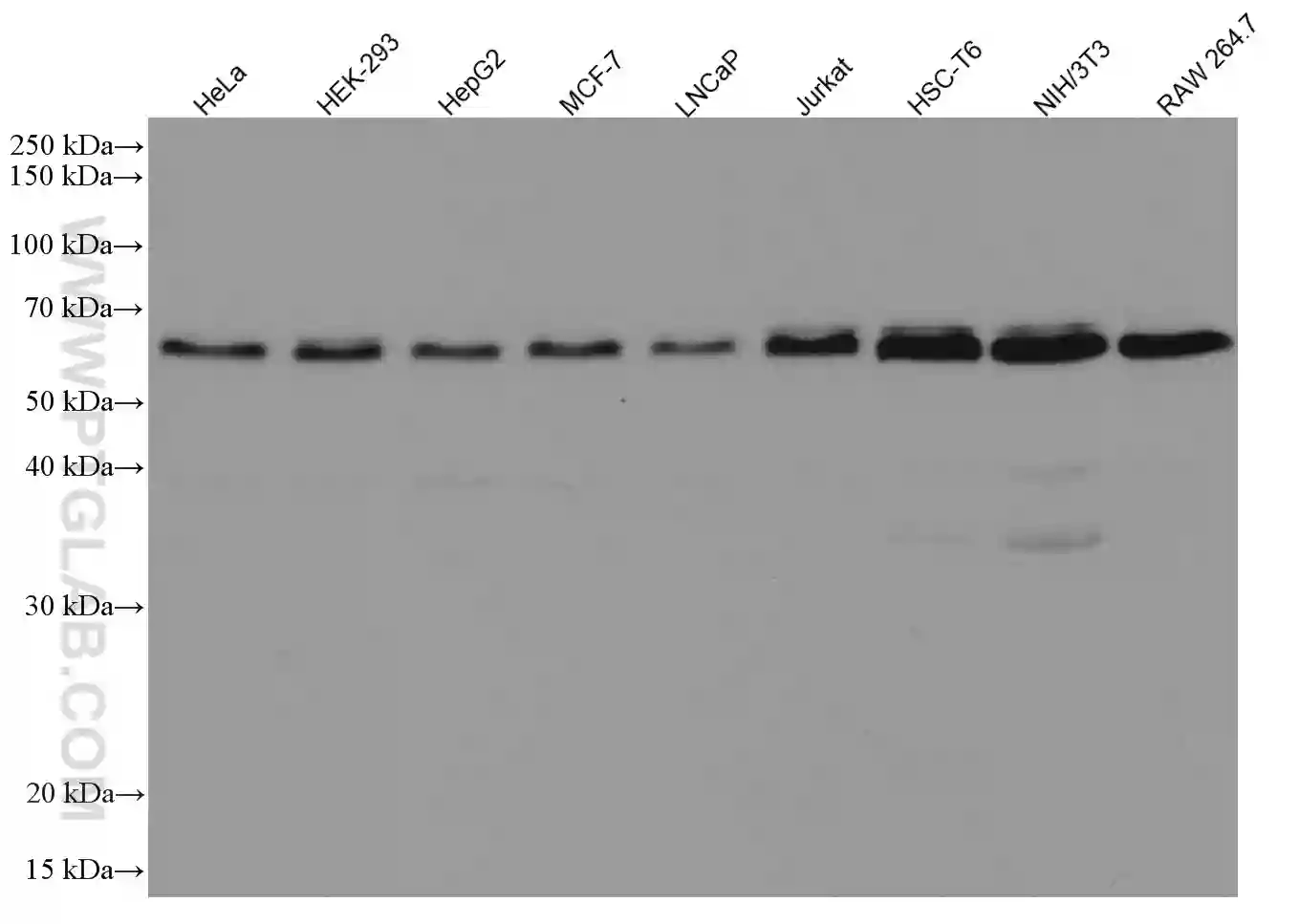
Western blot analysis of various lysates using monoclonal Beclin1 antibody (66665-1-Ig). |
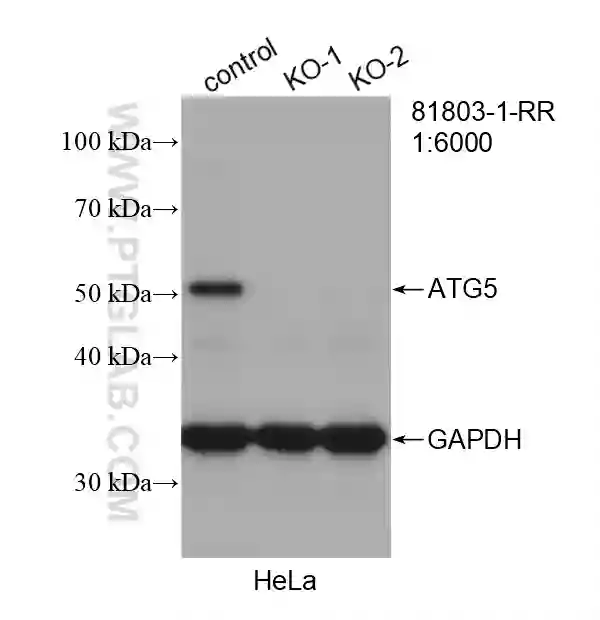
Western blot analysis of wild-type and ATG5 knockout Hela cells using recombinant ATG5 antibody (81803-1-RR). |
Autophagy as a Tumor Promoter
While autophagy acts as a tumor suppressor, it can also promote tumor growth in advanced cancers. Autophagy is activated in the central part of solid tumors that are nutrient-poor and always exist under hypoxic conditions. Autophagy supports high energy demands of tumors for continued cell proliferation by supplying metabolic substrates from the recycled intracellular components [9].
Hypoxia-inducible factor-1 alpha (HIF-1α) regulates cellular changes under hypoxic conditions. HIF-1α is capable of rapidly inducing cell survival response by engaging the autophagy mechanism, implicating AMPK-mTOR and unfolded protein response (UPR) pathways [10,11]. The activated autophagy pathway indirectly influences HIF-1α to promote glucose metabolism [12]. Here, autophagy contributes to sustained tumor survival by enhancing stress tolerance and delivering nutrients to prevent cytotoxicity.
Functional mitochondria sustain the viability of Rat sarcoma (Ras)-transformed cells. Autophagy is required to replenish the pool of healthy mitochondria for oxidative metabolism when nutrients are in short supply [13]. As a matter of course, Ras-mutated cells are observed to have a high basal level of autophagy associated with the development of lung, colon, and pancreatic cancers.
Cancer cells possess the innate ability to metastasize, the migration, invasion, and colonization of new organs via the bloodstream. To successfully metastasize, cancer cells must thrive without an extracellular matrix (ECM). When cancer cells detach from their primary site, the loss of ECM induces cell death, called anoikis. Substantial evidence shows that autophagy confers resistance to anoikis and enables the proliferation of ECM-detached cancer cells [14]. In support of this, it was reported that the inhibition of autophagy minimizes anoikis resistance and lung metastasis of hepatocellular carcinoma [15].
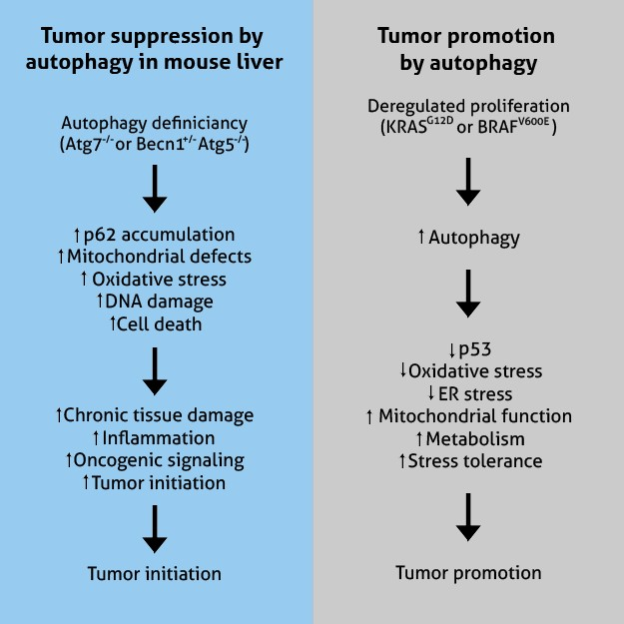
Figure adapted from White, 2015 (PMID: 25654549)
Therapeutic implications
One area of research that has generated much interest in recent years is the potential use of autophagy inhibitors and lysosomotropic agents such as chloroquine and hydroxychloroquine (HCQ) in cancer treatment. CQ and HCQ, combined with conventional chemotherapeutic agents, potentiate therapeutic activity by sensitizing cancer cells to various drugs. Besides autophagy inhibition, CQ and HCQ dysregulate Toll-like receptos9, p53, and CXCR4-CXCL12 pathways in cancer [16]. Hence, their potency and specificity remain outstanding concerns. Nonetheless, the quest for targeting autophagy has led to the development of class III lipid kinase, Vps34 inhibitors, that contributes to the early stages of autophagosome formation and ULK1/2 inhibitors with immense therapeutic promise [17].
Overall, the role of autophagy in cancer is complex and multifaceted. Understanding the mechanism by which autophagy affects cancer is an area of active research, and may hold the key to developing new therapies that target tumor microenvironments.
 |
 |
|
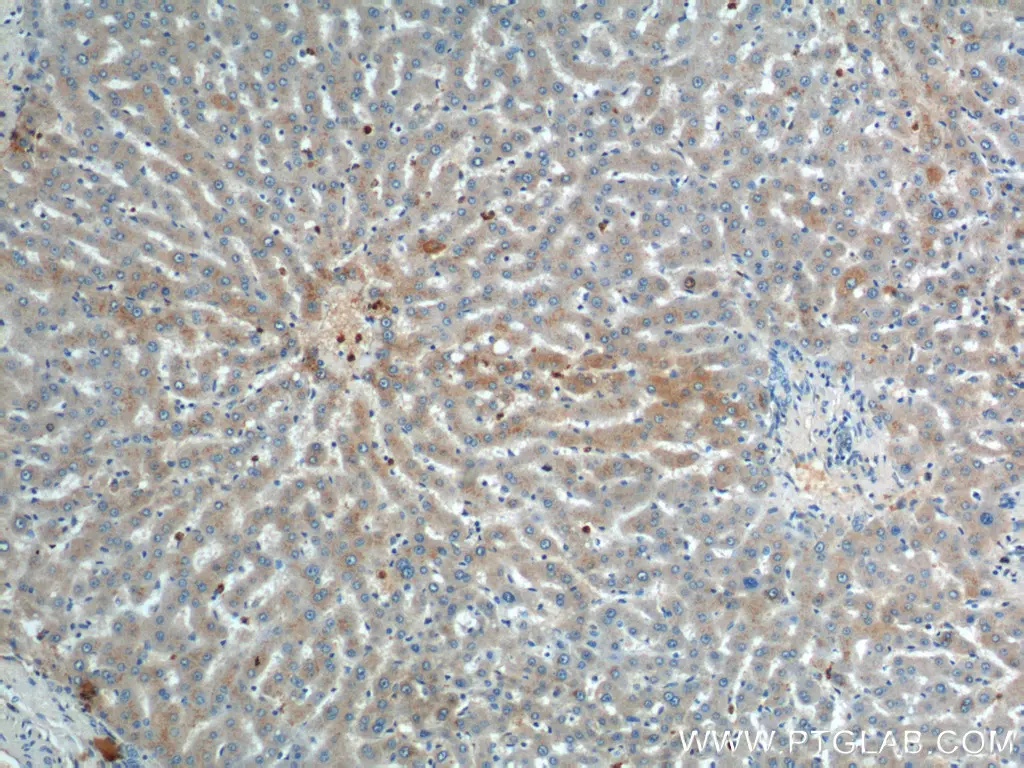
IHC analysis of human liver tissue using polyclonal ULK1 antibody (20986-1-AP). |
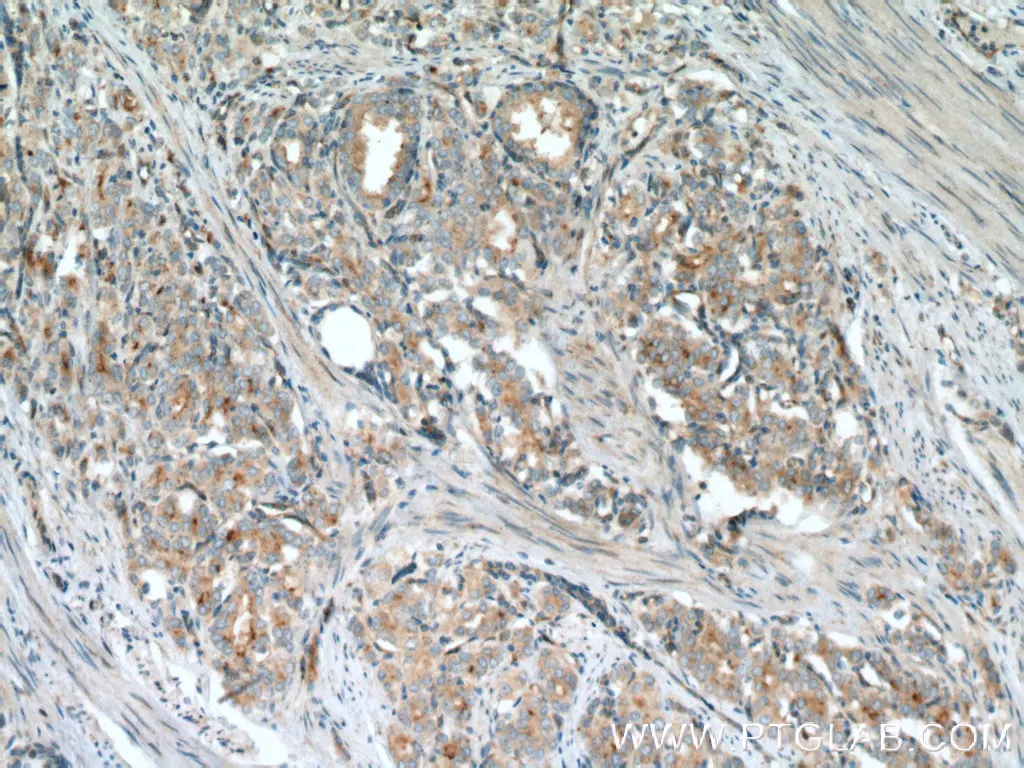
IHC analysis of human prostate cancer tissue using polyclonal VPS34 (C terminal) antibody (12452-1-AP). |
References
- Egan, D. F., Shackelford, D. B., Mihaylova, M. M., Gelino, S., Kohnz, R. A., Mair, W., ... Shaw, R. J. (2011). Phosphorylation of ULK1 (hATG1) by AMP-activated protein kinase connects energy sensing to mitophagy. Science (New York, N.Y.), 331(6016), 456–461. doi:10.1126/science.1196371
- Romanov, J., Walczak, M., Ibiricu, I., Schüchner, S., Ogris, E., Kraft, C., & Martens, S. (2012). Mechanism and functions of membrane binding by the Atg5-Atg12/Atg16 complex during autophagosome formation. The EMBO Journal, 31(22), 4304–4317. doi:10.1038/emboj.2012.278
- Fader, C. M., Sánchez, D. G., Mestre, M. B., & Colombo, M. I. (2009). TI-VAMP/VAMP7 and VAMP3/cellubrevin: two v-SNARE proteins involved in specific steps of the autophagy/multivesicular body pathways. Biochimica et Biophysica Acta, 1793(12), 1901–1916. doi:10.1016/j.bbamcr.2009.09.011
- Qu, X., Yu, J., Bhagat, G., Furuya, N., Hibshoosh, H., Troxel, A., ... Levine, B. (2003). Promotion of tumorigenesis by heterozygous disruption of the beclin 1 autophagy gene. The Journal of Clinical Investigation, 112(12), 1809–1820. doi:10.1172/JCI20039
- Liang, X. H., Jackson, S., Seaman, M., Brown, K., Kempkes, B., Hibshoosh, H., & Levine, B. (1999). Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature, 402(6762), 672–676. doi:10.1038/45257
- Morselli, E., Galluzzi, L., Kepp, O., Vicencio, J.-M., Criollo, A., Maiuri, M. C., & Kroemer, G. (2009). Anti- and pro-tumor functions of autophagy. Biochimica et Biophysica Acta, 1793(9), 1524–1532. doi:10.1016/j.bbamcr.2009.01.006
- Takamura, A., Komatsu, M., Hara, T., Sakamoto, A., Kishi, C., Waguri, S., ... Mizushima, N. (2011). Autophagy-deficient mice develop multiple liver tumors.Genes & Development, 25(8), 795–800. doi:10.1101/gad.2016211
- Moloney, J. N., & Cotter, T. G. (2018). ROS signalling in the biology of cancer. Seminars in Cell & Developmental Biology, 80, 50–64. doi:10.1016/j.semcdb.2017.05.023
- Rabinowitz, J. D., & White, E. (2010). Autophagy and metabolism. Science (New York, N.Y.), 330(6009), 1344–1348. doi:10.1126/science.1193497
- Mazure, N. M., & Pouysségur, J. (2010). Hypoxia-induced autophagy: cell death or cell survival? Current Opinion in Cell Biology, 22(2), 177–180. doi:10.1016/j.ceb.2009.11.015
- Sinha, I., Sakthivel, D., Olenchock, B. A., Kruse, C. R., Williams, J., Varon, D. E., ... Wagers, A. J. (2017). Prolyl hydroxylase domain-2 inhibition improves skeletal muscle regeneration in a male Murine model of obesity. Frontiers in Endocrinology, 8, 153. doi:10.3389/fendo.2017.00153
- Masoud, G. N., & Li, W. (2015). HIF-1α pathway: role, regulation and intervention for cancer therapy. Acta Pharmaceutica Sinica. B, 5(5), 378–389. doi:10.1016/j.apsb.2015.05.007
- Guo, J. Y., Chen, H.-Y., Mathew, R., Fan, J., Strohecker, A. M., Karsli-Uzunbas, G., ... White, E. (2011). Activated Ras requires autophagy to maintain oxidative metabolism and tumorigenesis. Genes & Development, 25(5), 460–470. doi:10.1101/gad.2016311
- Guadamillas, M. C., Cerezo, A., & Del Pozo, M. A. (2011). Overcoming anoikis--pathways to anchorage-independent growth in cancer. Journal of Cell Science, 124(Pt 19), 3189– 3197. doi:10.1242/jcs.072165
- Peng, Y.-F., Shi, Y.-H., Ding, Z.-B., Ke, A.-W., Gu, C.-Y., Hui, B., ... Fan, J. (2013). Autophagy inhibition suppresses pulmonary metastasis of HCC in mice via impairing anoikis resistance and colonization of HCC cells. Autophagy, 9(12), 2056–2068. doi:10.4161/auto.26398
- Verbaanderd, C., Maes, H., Schaaf, M. B., Sukhatme, V. P., Pantziarka, P., Sukhatme, V., ... Bouche, G. (2017). Repurposing Drugs in Oncology (ReDO)-chloroquine and hydroxychloroquine as anti-cancer agents. Ecancermedicalscience, 11, 781. doi:10.3332/ecancer.2017.781
- Amaravadi, R. K., Kimmelman, A. C., & Debnath, J. (2019). Targeting autophagy in cancer: Recent advances and future directions. Cancer Discovery, 9(9), 1167–1181. doi:10.1158/2159-8290.CD-19-0292
- Figure 2: Ref: https://www.jci.org/articles/view/73941
Related Content
Autophagy in the pathogenesis of disease
What is the difference between pyroptosis and apoptosis
Cancer stem cells as a key to cure cancer
Metabolic regulation of cell death
Support
Newsletter Signup
Stay up-to-date with our latest news and events. New to Proteintech? Get 10% off your first order when you sign up.
