Chromobodies for live cell imaging
A Chromobody is a small intracellular functional antibody. It consists of a VHH/Nanobody that is genetically fused to a fluorescent protein.
What’s a Chromobody?
Chromobodies® are fluorescent nanoprobes for real-time and live-cell imaging of intracellular endogenous proteins. A Chromobody is a small intracellular functional antibody. It consists of a VHH/Nanobody that is genetically fused to a fluorescent protein. Plasmids encoding for Chromobodies can be transiently transfected into cells and the Chromobody protein is intracellularly expressed.
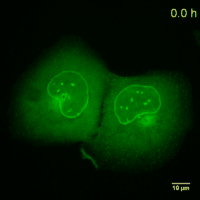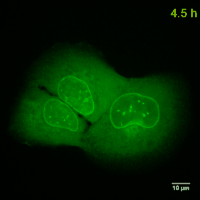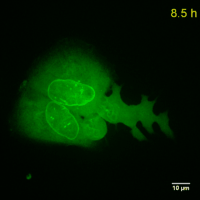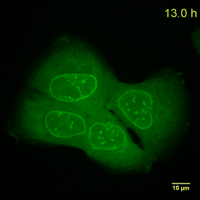
Why should you use a Chromobody?
Chromobodies non-invasively label their targets without interfering with target function. This makes them perfect nanoprobes for cellular research and high content analysis.
For example, the Actin-Chromobody has been thoroughly analyzed in comparison with multiple alternative Actin labeling methods and was ranked the least interfering probe for the detection of Actin (Actin visualization at a glance. Melak et al. J Cell Sci 2017).
Chromobodies don’t:
- affect cell viability or migration
- intercalate in DNA
- influence cellular functions
- show cytotoxic effects when overexpressing fluorescent fusion proteins
ChromoTek’s suite of seven Chromobodies are highlighted below, along with a list of published works utilizing Chromobodies® at the bottom of this blog.
Specificities of Chromobodies
Actin-Chromobody is a live-cell probe for visualization of the actin cytoskeleton and monitoring of its dynamics. The Actin-Chromobody enables non-invasive labeling of actin microfilaments not only in mammalian cells, but also in cells and tissues of evolutionary distant species such as Zebrafish and plants.
Cell Cycle-Chromobody visualizes the proliferating cell nuclear antigen (PCNA). PCNA is the homotrimeric ring-shaped protein that encircles the DNA and acts as a stationary loading platform for multiple, transiently interacting factors participating in various DNA transactions. The Cell Cycle-Chromobody enables you to screen compounds such as cancer drugs for effects on the cell cycle and toxicity in one experiment.
Dnmt1-Chromobody visualizes endogenous Dnmt1 in real-time during cell cycle progression. Dnmt1 1 is the major DNA methyltransferase in eukaryotic cells. It maintains genome-wide methylation patterns and plays an essential role in the epigenetic network controlling gene expression and genome stability during development.
Histone-Chromobody is a novel probe to visualize chromatin dynamics in live cells. It binds to the Histone H2A-H2B heterodimer which is part of the nucleosome core. The Histone-Chromobody has been tested in yeast, flies, and mammalian cells.
Lamin-Chromobody visualizes the nuclear lamina by binding to Lamin A/C. It enables you to observe cell size, nuclear morphology and mitosis in real time in your cell line of interest. Additionally, this Chromobody can be used for High-Content Analysis (HCA).
PARP1-Chromobody visualizes endogenous Poly (ADP-ribose) polymerase 1 (PARP1) in real time in live cells. PARP1 is one of the most abundant proteins in the nucleus and is involved in many cellular processes like DNA repair, transcriptional regulation, and modulation of chromatin structure. PARP1-Chromobody has been used to investigate DNA damage after microirradiation.
Vimentin-Chromobody enables tracing the dynamics of vimentin cytoskeleton in live cells. This Chromobody binds to Vimentin intermediate filaments, which are mainly present in mesenchymal cells.
Literature References by Chromobody
Actin-Chromobody
The Actin-Family Protein Arp4 Is a Novel Suppressor for the Formation and Functions of Nuclear F-Actin. (Yamazaki et al. Cell 2020) www.ncbi.nlm.nih.gov/pubmed/32204557
Characterization of 3D Printed Stretching Devices for Imaging Force Transmission in Live-Cells. (Mayer et al. Cell Mol Bioeng 2019) www.ncbi.nlm.nih.gov/pubmed/31719915
Actin chromobody imaging reveals sub-organellar actin dynamics. (Schiavon et al. bioRxiv 2019) www.biorxiv.org/content/10.1101/639278v3
Formin-2 drives polymerisation of actin filaments enabling segregation of apicoplasts and cytokinesis in Plasmodium falciparum. (Stortz et al. eLife 2019) www.ncbi.nlm.nih.gov/pubmed/31322501
Indirect visualization of endogenous nuclear actin by correlative light and electron microscopy (CLEM) using an actin‑directed chromobody. (Abdellatif et al. Histochem Cell Biol 2019) www.ncbi.nlm.nih.gov/pubmed/31154480
A Strategy to Optimize the Generation of Stable Chromobody Cell Lines for Visualization and Quantification of Endogenous Proteins in Living Cells. (Keller et al. Antibodies (Basel) 2019) www.ncbi.nlm.nih.gov/pubmed/31544816
A transient pool of nuclear F-actin at mitotic exit controls chromatin organization. (Baarlink et al. Nat Cell Biol 2017) www.ncbi.nlm.nih.gov/pubmed/29131140
Toxoplasma gondii F-actin forms an extensive filamentous network required for material exchange and parasite maturation. (Periz et al. Elife 2017) www.ncbi.nlm.nih.gov/pubmed/28322189
Actin visualization at a glance. (Melak et al. J Cell Sci 2017) www.ncbi.nlm.nih.gov/pubmed/28082420
Coordinate-targeted fluorescence nanoscopy with multiple off states. (Danzl et al. Nature Photonics 2016) www.nature.com/articles/nphoton.2015.266
Nuclear F-actin formation and reorganization upon cell spreading. (Plessner et al. J Biol Chem 2015) www.ncbi.nlm.nih.gov/pubmed/25759381
Live imaging of endogenous protein dynamics in zebrafish using chromobodies. (Panza et al. Development 2015) www.ncbi.nlm.nih.gov/pubmed/25968318
Fluorescent labelling of the actin cytoskeleton in plants using a cameloid antibody. (Rocchetti et al. Plant Methods 2014) www.ncbi.nlm.nih.gov/pubmed/24872838
Cell Cycle-Chromobody
The dynamic equilibrium of nascent and parental MCMs safeguards replicating genomes. (Sedlackova et al. bioRxiv 2019) www.biorxiv.org/content/10.1101/828954v1
A Multiplexed High-Content Screening Approach Using the Chromobody Technology to Identify Cell Cycle Modulators in Living Cells. (Schorpp et al. J Biomol Screen 2016) www.ncbi.nlm.nih.gov/pubmed/27044685
Nuclear actin modulates cell motility via transcriptional regulation of adhesive and cytoskeletal genes. (Sharili et al. Sci Rep 2016) www.ncbi.nlm.nih.gov/pubmed/27650314
Live imaging of endogenous protein dynamics in zebrafish using chromobodies. (Panza et al. Development 2015) www.ncbi.nlm.nih.gov/pubmed/25968318
Live cell imaging at the Munich ion microbeam SNAKE – a status report. (Drexler et al. Radiation Oncology 2015) www.ncbi.nlm.nih.gov/pubmed/25880907
Assessing kinetics from fixed cells reveals activation of the mitotic entry network at the S/G2 transition. (Akopyan et al. Mol Cell 2014) www.ncbi.nlm.nih.gov/pubmed/24582498
Quantitative live imaging of endogenous DNA replication in Mammalian cells. (Burgess et al. PLoS One 2012) www.ncbi.nlm.nih.gov/pubmed/23029203
Dnmt1-Chromobody
Long non-coding RNA PARTICLE bridges histone and DNA methylation. (O'Leary et al. Sci Rep 2017) www.ncbi.nlm.nih.gov/pubmed/28496150
Histone-Chromobody
Under the Microscope: Single-Domain Antibodies for Live-Cell Imaging and Super-Resolution Microscopy. (Traenkle & Rothbauer. Front Immunol 2017) www.ncbi.nlm.nih.gov/pubmed/28883823
Lamin-Chromobody
A Strategy to Optimize the Generation of Stable Chromobody Cell Lines for Visualization and Quantification of Endogenous Proteins in Living Cells. (Keller et al. Antibodies (Basel) 2019) www.ncbi.nlm.nih.gov/pubmed/31544816
Characterization of 3D Printed Stretching Devices for Imaging Force Transmission in Live-Cells. (Mayer et al. Cell Mol Bioeng 2019) www.ncbi.nlm.nih.gov/pubmed/31719915
A transient pool of nuclear F-actin at mitotic exit controls chromatin organization. (Baarlink et al. Nat Cell Biol 2017) www.ncbi.nlm.nih.gov/pubmed/29131140
Case study on live cell apoptosis-assay using lamin-chromobody cell-lines for high-content analysis. (Zolghadr et al. Methods Mol Biol 2012) www.ncbi.nlm.nih.gov/pubmed/22886277
Novel antibody derivatives for proteome and high-content analysis. (Schmidthals et al. Anal Bioanal Chem 2010) www.ncbi.nlm.nih.gov/pubmed/20372881
Targeting and tracing antigens in live cells with fluorescent nanobodies. (Rothbauer et al. Nat Methods 2006) www.ncbi.nlm.nih.gov/pubmed/17060912
PARP1-Chromobody
A New Nanobody-Based Biosensor to Study Endogenous PARP1 In Vitro and in Live Human Cells. (Buchfellner et al. PLoS One 2016) www.ncbi.nlm.nih.gov/pubmed/26950694
Vimentin-Chromobody
Real-time analysis of epithelial-mesenchymal transition using fluorescent single-domain antibodies. (Maier et al. Sci Rep 2015) www.ncbi.nlm.nih.gov/pubmed/26292717
Visualizing Epithelial-Mesenchymal Transition Using the Chromobody Technology. (Maier et al. Cancer Res 2016) www.ncbi.nlm.nih.gov/pubmed/27634766
Related Content
Nanobody-based reagents by ChromoTek
Intracellular Nanobodies for live-cell analysis in real-time
Actin Chromobody for live-cell super-resolution imaging
Visualize Histones in live cells: Histone-Chromobody
Visualize the onset of metastasis with Chromobodies
Actin Chromobody: Visualization of actin dynamics – NOT alteration
Novel cell cycle modulators identified using the Cell Cycle Chromobody Technology

Support
Newsletter Signup
Stay up-to-date with our latest news and events. New to Proteintech? Get 10% off your first order when you sign up.
