Post Translational Modifications: An Overview
Nature’s escape from genetic imprisonment
|
Navigate to section on page: |
A brief overview
What does PTM mean?
Cells need to detect and react to changes in internal and external conditions. One method used to adjust to these changes is chemical modification of proteins. Conditional chemical changes are relayed from sensors to effectors via reversible post-translational modifications (PTMs) of proteins. PTMs play an important part in modifying the end product of expression, contribute to biological processes and diseased conditions, playing a key role in many cellular processes such as cellular differentiation (1), protein degradation, signaling and regulatory processes, regulation of gene expression, and protein-protein interactions (2,3).
How does post translational modification work?
PTMs can happen at any step of the protein lifespan. Many proteins are modified shortly after translation is completed to mediate proper folding or to direct the nascent protein to distinct cellular locations (such as the nucleus or membrane). Other modifications occur after folding and localization are completed to activate or inactivate catalytic activity. Proteins are also covalently linked to tags that target a protein for degradation. They are modified through a combination of post-translational cleavage and the addition of functional groups through a step-wise mechanism of protein maturation or activation.
Where does post translational modification occur? PTMs occur at distinct amino acid side chains or peptide linkages and are most often mediated by enzymatic activity. Indeed, 5% of the proteome comprises enzymes that perform more than 200 types of PTMs (4). These enzymes include kinases, phosphatases, transferases, and ligases, which add or remove functional groups, proteins, lipids, or sugars to or from amino acid side chains, and proteases, which cleave peptide bonds to remove specific sequences or regulatory subunits. Many proteins can also modify themselves using autocatalytic domains, such as autokinase and autoprotolytic domains. PTMs can also be reversible based on the nature of the modification. As an example, phosphatases hydrolyze the phosphate group to remove it from the protein and reverse its biological activity (Figure 1).

Figure 1. Types of post-translational modifications (PTMs).
Most common post-translational modifications
Recent developments in mass-spectrometry (MS) methods have enabled the identification of thousands of PTM sites. Consequently, novel enrichment strategies have uncovered the global cellular importance of several types of modifications (e.g., acetylation, ubiquitylation, O-GlNac, N-linked glycosylation). More than 200 diverse types of PTMs are currently known (5,6), ranging from small chemical modifications (e.g., phosphorylation and acetylation) to the addition of complete proteins (e.g., ubiquitylation, Figure 3).
Phosphorylation
Protein phosphorylation (Figure 2) is the most commonly studied post-translational modification. It has been estimated that one-third of mammalian proteins may be phosphorylated, and this modification often plays a key role in modulating protein function. Phosphorylation takes place on serine, threonine, and tyrosine residues, acting to regulate protein function, enzymatic activity, protein–protein interactions, and protein localization. Phosphorylation is catalyzed by kinases and can be reversible – phosphorylated proteins can be dephosphorylated by phosphatases.

Figure 2. WB result of phospho-Marcks antibody (10018-3-AP, 1:1500) with mouse J774 macrophage cells treated with PMA.
Glycosylation and Glycanation
The majority of proteins that are synthetized on ribosomes associated with the endoplasmic reticulum undergo glycosylation. That means a covalent attachment of sugar moieties is added to the polypeptide chain. The two most common types of glycosylation in Eukaryotes are N-linked glycosylation – to asparagine, and O-linked glycosylation – to serine and threonine. Glycosidase enzymes can be incorporated into your Western blot sample preparation to identify the presence of glycosylation.
Ubiquitination
Protein ubiquitination means a covalent ubiquitin is added to lysine, cysteine, serine, threonine, or directly to the protein N-terminus. Ubiquitin is a small (+/-8.6 kDa) protein expressed across almost all tissue types (Figure 3). Ubiquitination is an enzymatic reaction catalyzed by a three-enzyme cascade (E1, E2, and E3). That provides substrate specificity and activation, conjugation, and ligation steps. Proteins can be monoubiquitinated (with one ubiquitin molecule) or polyubiquitinated. Polyubiquitination takes place when additional ubiquitin molecules are added to the initial ubiquitin molecule. Ubiquitination via the proteome can mark proteins for degradation. It is also important for cellular signaling, the internalization of membrane proteins , and the development and regulation of transcription.
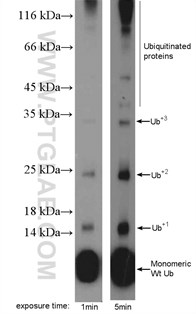
Figure 3. MDA-MB-453s cells were subjected to SDS PAGE followed by western blot with 10201-2-AP (ubiquitin antibody) at a dilution of 1:600.
PTMs impact on health and disease
The analysis of proteins and their PTMs is particularly important for the study of heart disease, cancer, neurodegenerative diseases, and diabetes (7). The main challenges in studying post-translationally modified proteins are the development of specific detection and purification methods. Fortunately, these technical obstacles are being overcome with a variety of new and refined proteomics technologies.
References:
- Chemical biology: dressed-up proteins.
- Concepts in sumoylation: a decade on.
- Proteomic analysis in the neurosciences.
- The Roles of Post-translational Modifications in the Context of Protein Interaction Networks
- Deciphering a global network of functionally associated post-translational modifications.
- PTMcode: a database of known and predicted functional associations between post-translational modifications in proteins.
- Citrullination: a posttranslational modification in health and disease.
Related Products
Western blot products overview
Tag & loading control antibodies
SignalBright chemiluminescent substrate
Related Content
Detecting low abundance proteins via Western blot
In search of low molecular weight proteins
Lysate preparation: why RIPA buffer is best
Lysate preparation: how to optimize your extraction
Why does the observed protein molecular weight (MW) differ from the calculated one?
Proteases for Western blotting: Choosing the right tools for the job
Loading controls for Western blotting
Support
Newsletter Signup
Stay up-to-date with our latest news and events. New to Proteintech? Get 10% off your first order when you sign up.


