How to elute GFP-fusion protein from the GFP-Trap®
Tips and tricks
Binding of GFP-fusion proteins to the ChromoTek GFP-Trap® is very strong (this Nano-Trap has a dissociation constant in the pico molar range) and allows stringent washing, because it is stable up to 70°C and functional active in up to:
- 2M NaCl
- 8M Urea
- 0.2% SDS
That is beneficial for reducing the background noise from nonspecific binding and provides you cleaner extracts for your downstream applications. It really makes the GFP-Trap® a perfect tool for reproducible, fast and clean immunoprecipitations.
But how to elute the GFP-fusion protein from the GFP-Trap®?
The strong binding of GFP-fusion proteins to the GFP-Trap makes its elution a bit challenging. Let me explain this: An elution of the bound GFP-fusion protein by a competitive peptide, which matches the Trap’s epitope, and which replaces the GFP-fusion protein doesn’t work. This is due to the nature of nanobodies, which bind 3-dimensional epitopes rather than (linear) peptide sequences. Also the addition of chaotropic compounds like urea don’t elute the bond GFP-fusion protein as the GFP-Trap® works under denaturing conditions.
Now, what can you do?
A very effective way to elute your GFP-tagged protein is SDS sample buffer. That always works and results in denatured GFP-fusion protein. Alternatively you may elute with glycine at pH 2.5 (incubation time: 30 sec under constant mixing). However the fusion protein may effect this elution strategy: As residual fusion protein may stick to the agarose beads, you may repeat the glycine elution to improve elution. Very important: Don’t forget to neutralize proteins immediately after elution!
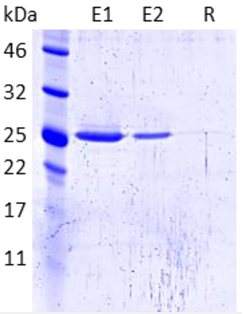
Immunoprecipitation of EGFP (28.0 kDa) using GFP-Trap Agarose and elution with Glycine elution buffer.
What other options do I have?
You may consider to introduce a protease cleavage site between GFP and the fusion protein. This option is recommended if you are dealing with less stable proteins.
Last but not least, you should always check if you really need to elute the GFP-fusion protein at all. Many experiments can be conducted using the bound GFP-fusion protein:
- Proteins can be digested when still coupled to the beads for subsequent mass spectrometry analysis:
On-bead digestion protocol: From immunoprecipitation with nanobodies to MS analysis - Enzymatic activity assays can be performed when still coupled to the beads if the active center is not blocked.
Related Content
What GFP-Trap should I use for my immunoprecipitation?
GFP-Trap for immunoprecipitation (IP) – stringent washing
Mass spec-compatible immunoprecipiation for GFP, mNeonGreen, Myc, RFP, Spot, and TurboGFP
GFP (green fluorescent protein): Properties, origin, specifications, tips
The best anti-GFP antibody for immunoprecipitation: GFP-Trap
Green fluorescent protein (GFP) in plant research
GFP-Booster: GFP Nanobody for better images in immunofluorescence
GFP and RFP-Booster for better immunofluorescence imaging
GFP- Nanobodies and RFP- Nanobodies conjugated to quantum dots
Learn how to save precious hours on your IP, IF, and western blotting experiments

Support
Newsletter Signup
Stay up-to-date with our latest news and events. New to Proteintech? Get 10% off your first order when you sign up.