Tested Applications
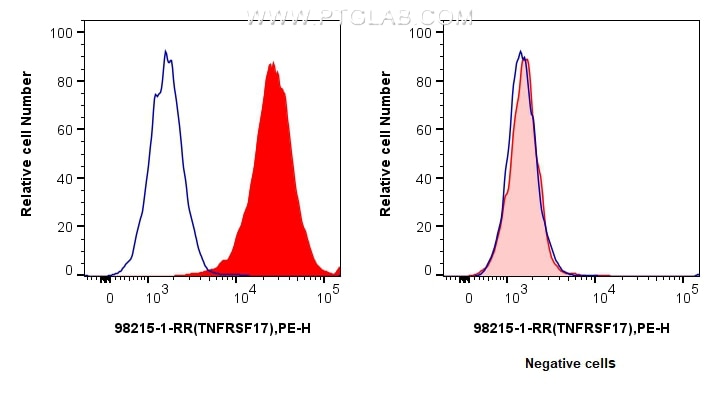
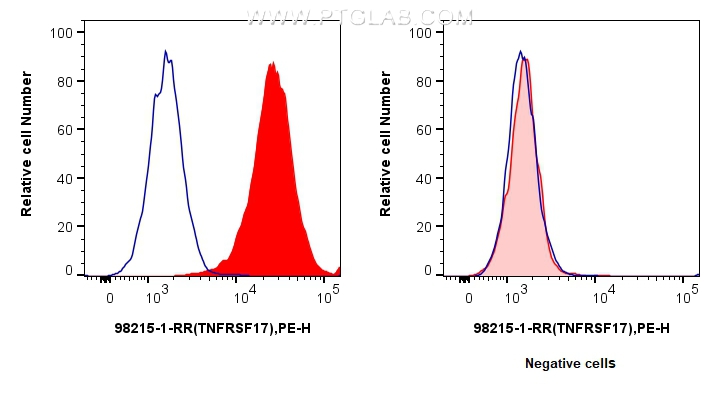
| Positive FC detected in | U266 cells |
Recommended dilution
| Application | Dilution |
|---|---|
| This reagent has been tested for flow cytometric analysis. It is recommended that this reagent should be titrated in each testing system to obtain optimal results. | |
| Sample-dependent, Check data in validation data gallery. | |
Product Information
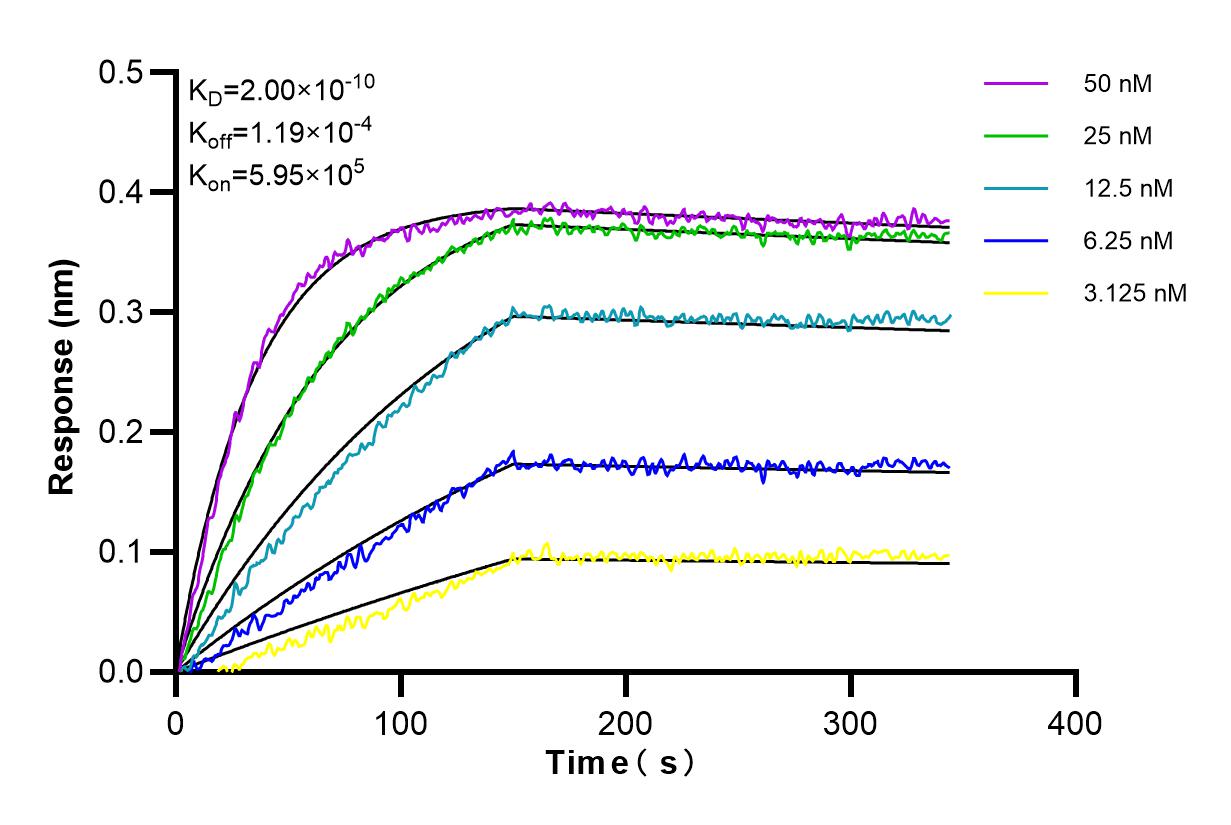
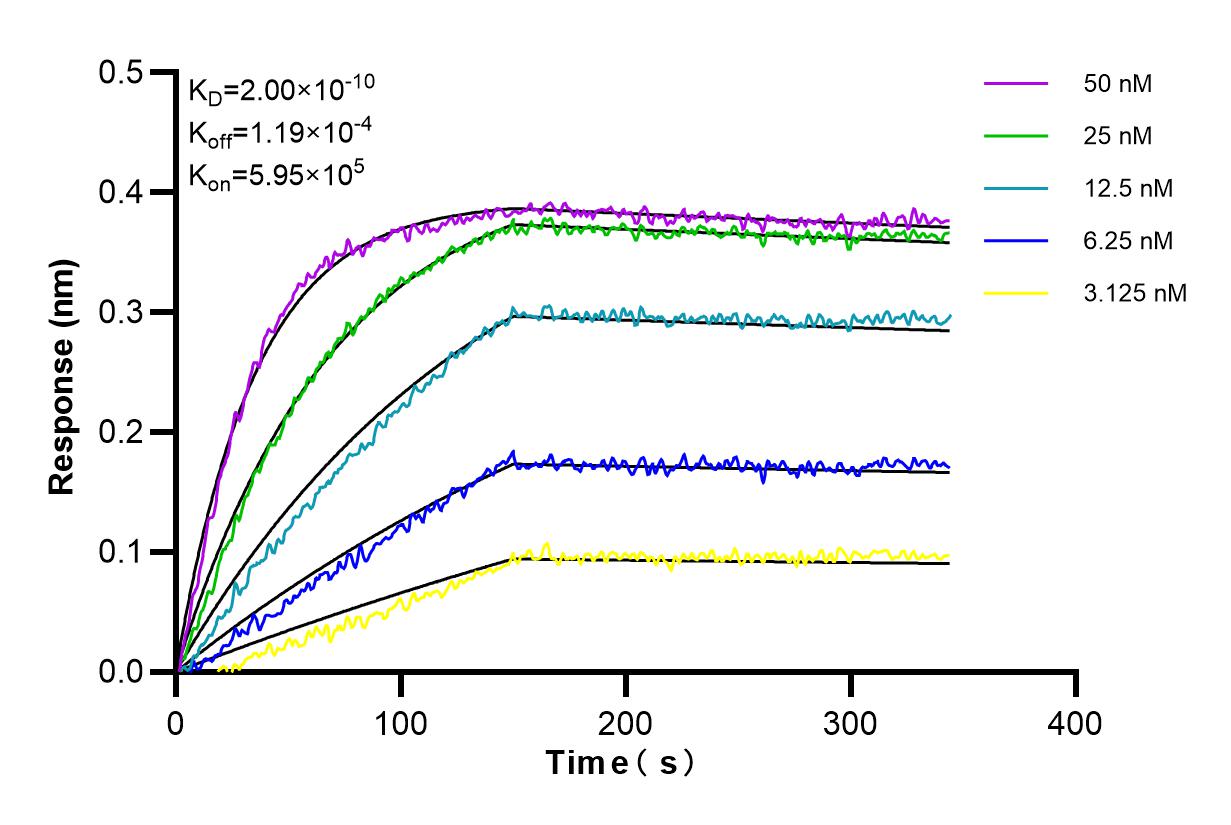
98215-1-RR targets BCMA/TNFRSF17 in FC applications and shows reactivity with human samples.
| Tested Reactivity | human |
| Host / Isotype | Rabbit / IgG |
| Class | Recombinant |
| Type | Antibody |
| Immunogen | Recombinant protein Predict reactive species |
| Full Name | tumor necrosis factor receptor superfamily, member 17 |
| Calculated Molecular Weight | 184 aa, 20 kDa |
| GenBank Accession Number | BC058291 |
| Gene Symbol | TNFRSF17 |
| Gene ID (NCBI) | 608 |
| Conjugate | Unconjugated |
| Form | Liquid |
| Purification Method | Protein A purfication |
| UNIPROT ID | Q02223 |
| Storage Buffer | PBS with 0.09% sodium azide, pH 7.3. |
| Storage Conditions | Store at 2 - 8°C. Stable for one year after shipment. |
Background Information
BCMA (B cell maturation antigen), also known as TNFRSF17, is 20.2-kDa type III transmembrane glycoprotein and is a member of the TNF-receptor superfamily. This receptor is preferentially expressed in mature B lymphocytes and plasma cells, which may be important for B cell development and autoimmune response (PMID: 32943087; 9846698). BCMA has two agonist ligands: a proliferation-inducing ligand (APRIL) and BAFF. When BCMA binds to APRIL, it transmits signals of cell survival and proliferation (PMID: 27127303); when BCMA binds to BAFF, it mediates the activation of NF-kappaB and MAPK8/JNK (PMID: 36140254). It has been found that the overexpression and activation of BCMA are associated with multiple myeloma (MM) in preclinical models and humans, supporting its potential utility as a therapeutic target for MM (PMID: 32055000; 32943087).
Protocols
| Product Specific Protocols | |
|---|---|
| FC protocol for BCMA/TNFRSF17 antibody 98215-1-RR | Download protocol |
| Standard Protocols | |
|---|---|
| Click here to view our Standard Protocols |