The importance of epitope selection in experimental design
Discover how pinpointing the right antibody binding site can transform the reliability and impact of your research, diagnostics, and therapeutic strategies.
When designing an experiment that uses antibodies, one often overlooked parameter is the exact binding site of your antibody on the protein of interest, also known as the epitope. An antibody’s primary role is to bind to a specific protein (also known as the antigen or immunogen) which in turn allows for the detection, neutralization, or modification of the protein’s activity. Carefully choosing your antibodies based on epitope binding site can greatly influence experimental outcomes, therapeutic strategies, and diagnostic accuracy. In this blog we will walk through 10 examples where having more knowledge on your antibody’s epitope binding site is critical.
Conformational vs. Linear Epitopes
An antibody that binds a linear epitope may work in Western blot but fail in immunohistochemistry (IHC), where protein conformation remains intact. Different antibody applications may have different criteria for successful binding of antibodies to epitopes. Some assays, like ELISA, could require either conformational or linear epitopes depending on the sample preparation. Experiments such as Immunoprecipitation (IP) or Mass-Spectrometry often digest or denature proteins before analysis, which would present linear epitopes.
Detecting Post-Translational Modifications
If an epitope is near a phosphorylation, glycosylation, or acetylation site, modifications may alter binding, affecting experimental results. Alternatively, you may want an antibody that binds selectively to a post-translational modification, such as a specific phosphorylation site.

^ Non-treated and Calyculin A treated HEK-293 cells were subjected to SDS PAGE followed by western blot with 80455-1-RR (Phospho-AKT (Ser473) antibody) at dilution of 1:10000 incubated at room temperature for 1.5 hours. The membrane was stripped and re-blotted with GAPDH antibody as loading control.
Differentiation of Isoforms
Antibodies targeting epitopes unique to specific isoforms help distinguish between closely related proteins in experiments. One example of this is the ability to differentiate between PKM1 and PKM2, two isoforms of pyruvate kinase that play distinct roles in cellular metabolism. PKM1 is found in muscle and brain while PKM2 is found in cancer cells and rapidly proliferating tissues. One isoform is important for efficient and healthy energy production and the other is linked to metabolic reprogramming in cancer (the Warburg effect).

^ Mouse skeletal muscle tissue was subjected to SDS PAGE followed by western blot with 15821-1-AP (PKM1-specific antibody) at dilution of 1:2000, incubated at room temperature for 1.5 hours.
Steric Hindrance and Interference
In multiplex assays like flow cytometry or mass spectrometry where multiple antibodies are used to detect different proteins simultaneously, knowing the epitopes is vital for preventing interference. By selecting antibodies that target non-overlapping epitopes, researchers can ensure that each antibody binds specifically to its target, allowing for clearer and more accurate data. In flow cytometry applications, cell activation sites may also be useful for detection. Ensuring the use of antibodies targeted to different epitopes of that protein is critical for the success of such an assay.
If an antibody’s epitope is buried in a protein complex, the antibody targeting that epitope may not bind effectively in co-immunoprecipitation or pull-down assays. Using well-validated antibodies for IP and Co-IP is a great start but knowing more about your antibody’s epitope binding region can also provide some insight into whether an antibody is a good fit for your experiment or not.
Epitope Masking by Protein Folding or Cellular Structure
In live-cell imaging or flow cytometry, antibodies may fail to bind if the epitope is inaccessible on the native protein surface. It is important for experimental design to understand whether your antibody binds to the intracellular or extracellular portion of transmembrane proteins. Intracellular epitopes would require fixation and permeabilization, but extracellular epitopes would not.
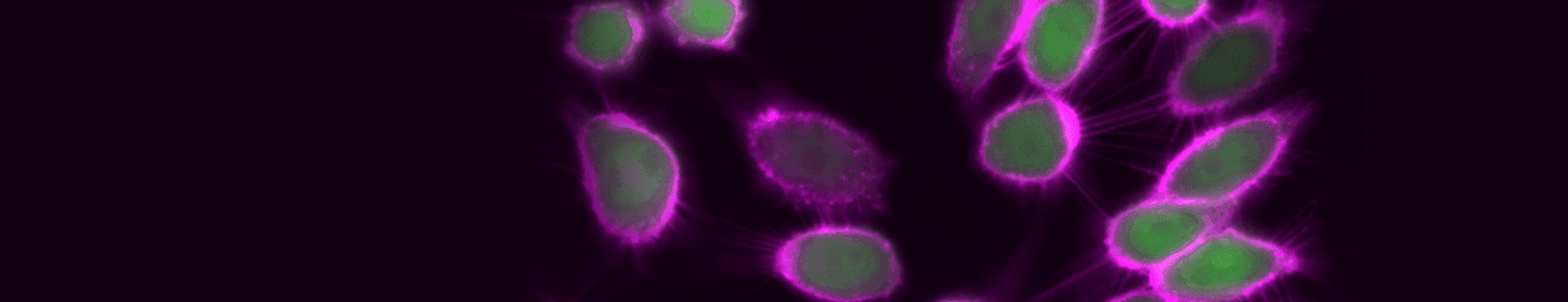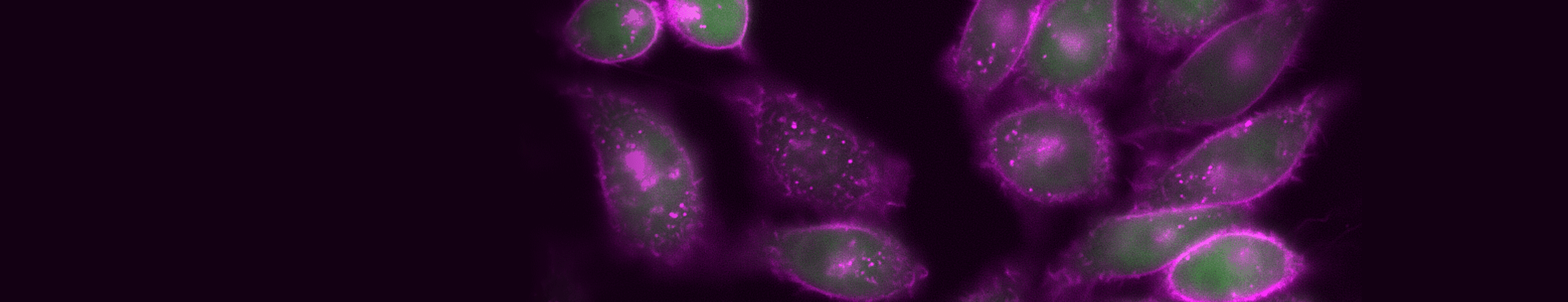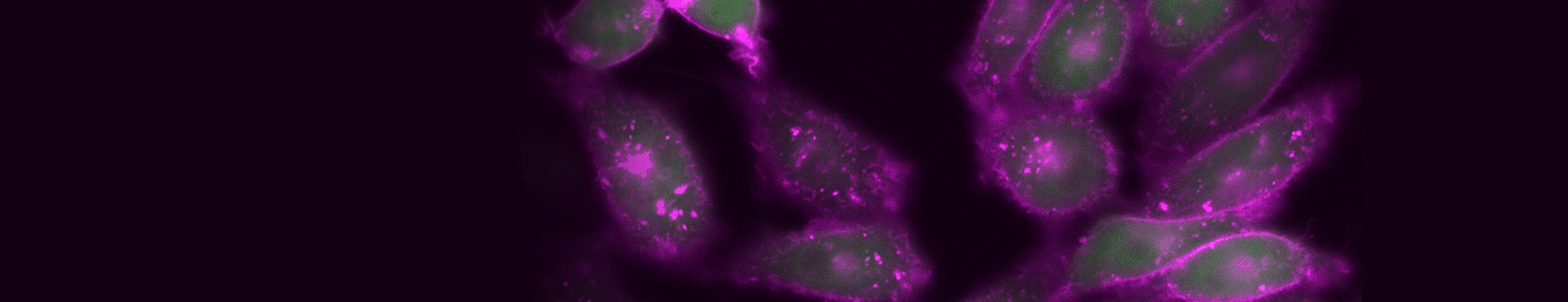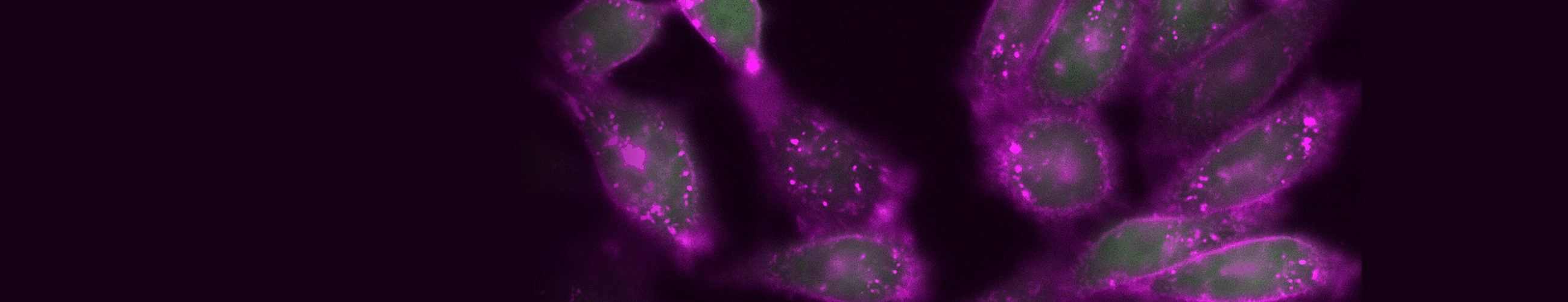
Time-lapse imaging of TIGIT internalization, detected with CoraLite® Plus 647-conjugated TIGIT VHH (magenta).
Subcellular localization is also important to consider. If you want to investigate a protein’s role in the nucleus, cytoplasm, or membrane, it is important to choose an antibody that recognizes an epitope exposed in that compartment. This ensures that the antibody accurately reflects the protein’s true localization, which is critical for experiments such as immunofluorescence, microscopy, or immunocytochemistry.
Proteolytic Cleavage Detection
Antibodies targeting cleavage sites can track protein processing events, such as caspase cleavage in apoptosis studies. PARP1 is another protein involved in DNA damage where the detection of PARP fragments is commonly considered an important biomarker of apoptosis. Proteolytic cleavage is a fundamental regulatory mechanism of other biological functions such as hormone precursors, blood coagulation cascade, complement proteins in immune activation, Notch receptor signaling, and viral entry and pathogenesis, just to name a few.

^ Various lysates were subjected to SDS PAGE followed by western blot with 25128-1-AP (cleaved Caspase 3 antibody) at dilution of 1:1000 incubated at room temperature for 1.5 hours.
Identifying Protein Degradation
Similar to proteolytic cleavage, protein degradation can lead to some confusing or unexpected results in experiments. For example, actin can be degraded by many different proteases and biological events like apoptosis. The most familiar form of actin (42kDa) will produce typical fragments including 35-38 kDa, ~30 kDa, and 15 kDa after degradation. The 35-38 kDa fragment is most common due to enzymatic cleavage between Val43 and Met44 amino acids.
Understanding whether your antibody identifies either full-length or fragmented actin will help you understand the results of your western blot or experiment. Proteintech’s 60008-1-Ig Beta Actin Monoclonal Antibody can recognize a degraded fragment but 66009-1-Ig Beta Actin Monoclonal Antibody will only detect the full-length protein because its binding site is located in the first 43 aa. Using epitope binding region to select your antibody can help achieve accurate and precise results by targeting the appropriate region of the protein.

^ Western blot analysis of Beta-actin in various tissues and cell lines using Proteintech antibody 60008-1-Ig at a dilution of 1:5000. Extra bands were detected in some species with unknown reason.
Therapeutic Targeting in Drug Development
Epitope location affects whether an antibody blocks receptor-ligand interactions or activates/inhibits signaling pathways in therapeutic research. Blocking and neutralizing antibodies are examples of these biological functions. In the case of viral infections, neutralizing antibodies often target specific epitopes on viral proteins to prevent the virus from entering host cells. Identifying these epitopes is imperative for developing an effective treatment.

^ Recombinant human IL-6 (Cat.NO. HZ-1019) stimulates proliferation of hybridoma (Proteintech mouse anti-GST clone 3G12B10) in a dose-dependent manner (blue curve, refer to bottom X-left Y axis). The activity of human IL-6 (1 ng/mL HZ-1019) is neutralized by mouse anti-human IL-6 monoclonal antibody 69001-1-Ig at serial dose (red curve, refer to top X-right Y axis). The ND50 is typically 10-30 ng/mL.
Epitope mapping is particularly important in the development of therapeutic antibodies, such as those used for cancer therapy or autoimmune diseases. Understanding which region of a target protein an antibody binds to can have a significant impact on its therapeutic effectiveness.
In oncology, monoclonal antibodies can be used to target cancer cell surface antigens. Understanding the epitope allows for the development of antibodies that bind to regions critical for the cancer cell’s growth or survival, improving the efficacy of treatments and minimizing off-target effects.
Antibody Pair Compatibility
Understanding the specific epitopes that antibodies recognize is fundamental to the design and optimization of immunoassays, including Enzyme-Linked Immunosorbent Assays (ELISA). In immunoassays such as sandwich ELISAs, antibody pairs are used. Antibody pairs include a capture antibody and a detection antibody that bind to different non-overlapping epitopes on the same protein, which increases the sensitivity and specificity of the assay. The sandwich ELISA assay approach is used in multiplex platforms like Luminex, Cisbio, Meso Scale Diagnostics, AlphaLISA assays, fluorescence resonance energy transfer (FRET), and Proximity Ligation Assays (PLA), among others. Assay developers typically test many antibodies to find the ideal pair and may also utilize techniques such as epitope binning. Therefore, information on antibody epitope binding sites is tremendously helpful for multiplex assay design.
In conclusion, the epitope of an antibody is a fundamental factor that determines how effectively the antibody performs in various applications. From ensuring specificity and reducing cross-reactivity to optimizing experimental designs and enhancing therapeutic outcomes, understanding and mapping the epitope is crucial for improving the reliability, accuracy, and effectiveness of antibody-based tools. Whether in research, diagnostics, or therapeutics, knowing where an antibody binds on its target protein is indispensable for achieving precise, reproducible, and meaningful results. Therefore, characterizing the epitope is not just a technical step—it is essential for the success of any experiment that relies on antibodies.
Related Content
Blog: Every protein tells a story | Here is why every unique epitope matters
Support
Newsletter Signup
Stay up-to-date with our latest news and events. New to Proteintech? Get 10% off your first order when you sign up.

