Validation Data Gallery
Product Information
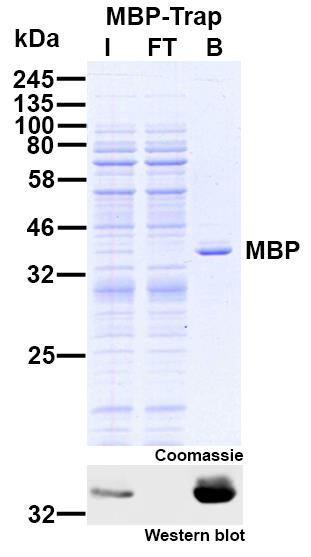
The ChromoTek MBP-Trap Agarose are affinity beads for IP of MBP-fusion proteins. They comprise MBP Nanobodies/ VHHs coupled to agarose beads.
| Description | Immunoprecipitation of MBP-fusion proteins and their interacting factors with anti-MBP Nanobody conjugated to beads.
• Immunoprecipitation of low abundant MBP-fusion proteins • Significantly higher sensitivity than amylose resin and other affinity resins • No heavy & light antibody chains • Stable under harsh washing conditions • Suitable for downstream mass spec analysis |
| Applications | IP, CoIP, ChIP, RIP |
| Specificity/Target | Maltose-binding protein (MBP) of Escherichia coli |
| Binding capacity | 45 μg (0.625 nmol) of recombinant MBP-tagged protein (~74.2 kDa) per 25 μL bead slurry |
| Conjugate | Agarose beads; bead size: ~ 90 µm (cross-linked 4 % agarose beads) |
| Elution buffer | SDS sample buffer 0.2 M glycine pH 2.5 |
| Wash buffer compatibility | 2 M urea, 2 M NaCl, 10 mM DTT, 2 % Nonidet P40 Substitute, 1 % Triton X-100 |
| Type | Nanobody |
| Class | Recombinant |
| Host | Alpaca |
| Affinity (KD) | Dissociation constant KD of 4 nM |
| Compatibility with mass spectrometry | TheMBP-Trap is optimized for on-bead digestion. For the application note, please click here: On-bead digest protocol for mass spectrometry |
| RRID | AB_2631382 |
| Storage Buffer | 20% EtOH |
| Storage Condition | Upon receipt store at +4°C. Do not freeze! |
| Size | 25ul/reactions (eg:20rxns=500ul slurry) |
Documentation
| SDS |
|---|
| mbta_SDS_MBP-Trap Agarose (EN) |
| Datasheet |
|---|
| MBP-Trap Agarose beads Datasheet |
| Whitepaper |
|---|
| Immunoprecipitation of GST- and MBP-fusion proteins (PDF) |
| Trouble shooting |
|---|
| Troubleshooting guide immunoprecipitation (PDF) |
Publications
| Application | Title |
|---|---|
Mol Cell A PARylation-phosphorylation cascade promotes TOPBP1 loading and RPA-RAD51 exchange in homologous recombination. | |
Plant Cell FRA1 Kinesin Modulates the Lateral Stability of Cortical Microtubules through Cellulose Synthase-Microtubule Uncoupling Proteins. | |
Plant Cell TTL Proteins Scaffold Brassinosteroid Signaling Components at the Plasma Membrane to Optimize Signal Transduction in Arabidopsis. | |
Curr Biol Arabidopsis pavement cell shape formation involves spatially confined ROPGAP regulators. |
Reviews
The reviews below have been submitted by verified Proteintech customers who received an incentive for providing their feedback.
FH Daniel (Verified Customer) (09-05-2025) | I successfully used these beads to capture recombinant MBP-tagged protein and perform in vitro phosphorylation assay with MBP-tagged protein on beads. Much lower background than other beads.
|