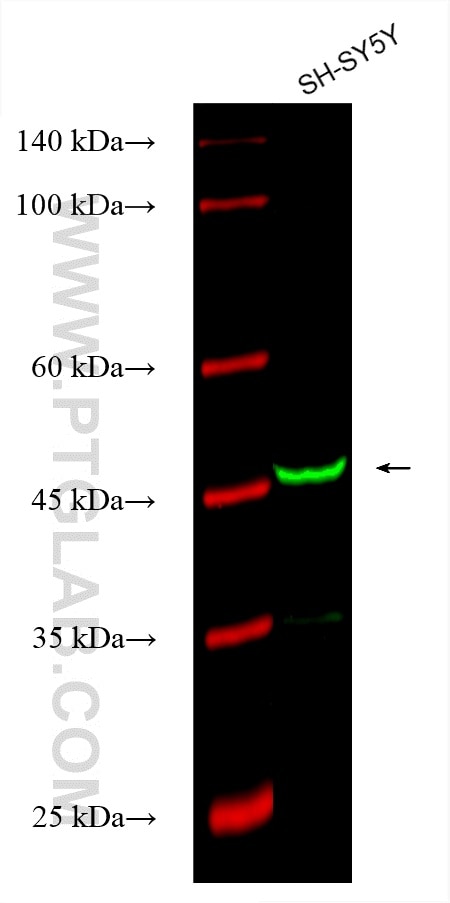
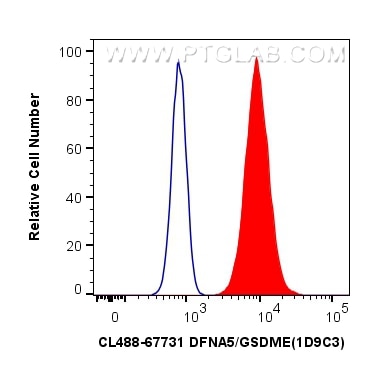
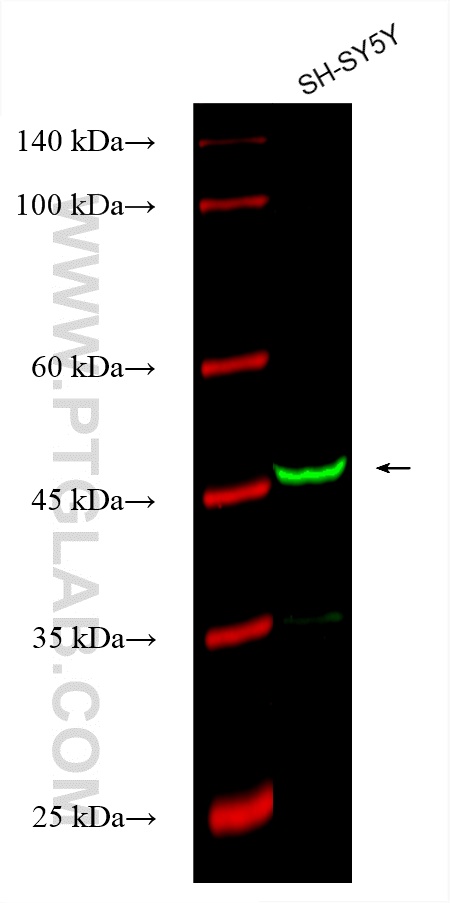
Tested Applications
| Positive WB detected in | SH-SY5Y cells |
| Positive FC (Intra) detected in | SH-SY5Y cells |
Recommended dilution
| Application | Dilution |
|---|---|
| Western Blot (WB) | WB : 1:500-1:1000 |
| Flow Cytometry (FC) (INTRA) | FC (INTRA) : 0.40 ug per 10^6 cells in a 100 µl suspension |
| It is recommended that this reagent should be titrated in each testing system to obtain optimal results. | |
| Sample-dependent, Check data in validation data gallery. | |
Product Information
CL488-67731 targets DFNA5/GSDME in WB, FC (Intra) applications and shows reactivity with human, mouse, rat, pig samples.
| Tested Reactivity | human, mouse, rat, pig |
| Host / Isotype | Mouse / IgG1 |
| Class | Monoclonal |
| Type | Antibody |
| Immunogen | DFNA5/GSDME fusion protein Ag30514 Predict reactive species |
| Full Name | deafness, autosomal dominant 5 |
| Calculated Molecular Weight | 496 aa, 55 kDa |
| Observed Molecular Weight | 50-55 kDa |
| GenBank Accession Number | BC019689 |
| Gene Symbol | DFNA5 |
| Gene ID (NCBI) | 1687 |
| RRID | AB_3084376 |
| Conjugate | CoraLite® Plus 488 Fluorescent Dye |
| Excitation/Emission Maxima Wavelengths | 493 nm / 522 nm |
| Form | Liquid |
| Purification Method | Protein G purification |
| UNIPROT ID | O60443 |
| Storage Buffer | PBS with 50% glycerol, 0.05% Proclin300, 0.5% BSA, pH 7.3. |
| Storage Conditions | Store at -20°C. Avoid exposure to light. Stable for one year after shipment. Aliquoting is unnecessary for -20oC storage. |
Background Information
DFNA5 (deafness, autosomal dominant 5), also known as GSDME or ICERE-1, is a 496 amino acid protein that is expressed in cochlea tissue, as well as in placenta, brain, heart, liver, lung and pancreas. Defects in the gene encoding DFNA5 are the cause of non-syndromic sensorineural deafness autosomal dominant type 5 (DFNA5), a form of sensorineural hearing loss that results from damage to one of various structures that receive sound information in the brain.
Protocols
| Product Specific Protocols | |
|---|---|
| WB protocol for CL Plus 488 DFNA5/GSDME antibody CL488-67731 | Download protocol |
| FC protocol for CL Plus 488 DFNA5/GSDME antibody CL488-67731 | Download protocol |
| Standard Protocols | |
|---|---|
| Click here to view our Standard Protocols |