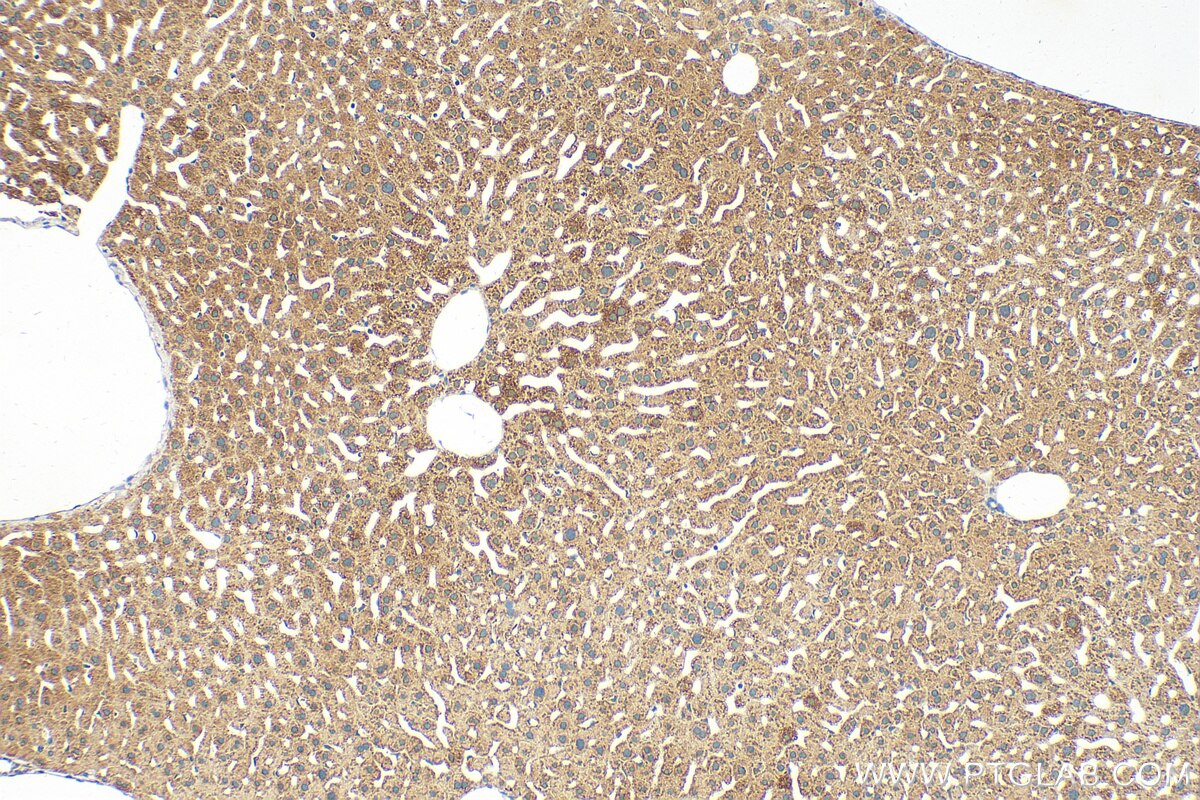
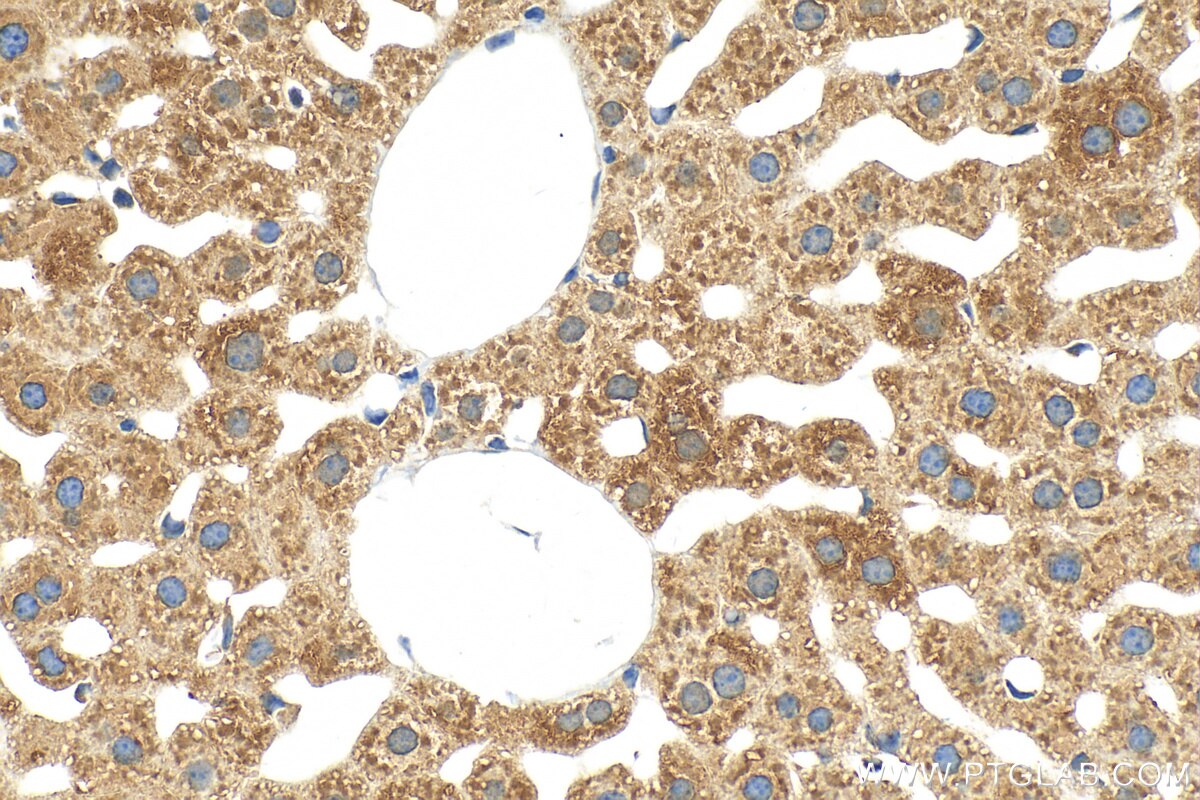
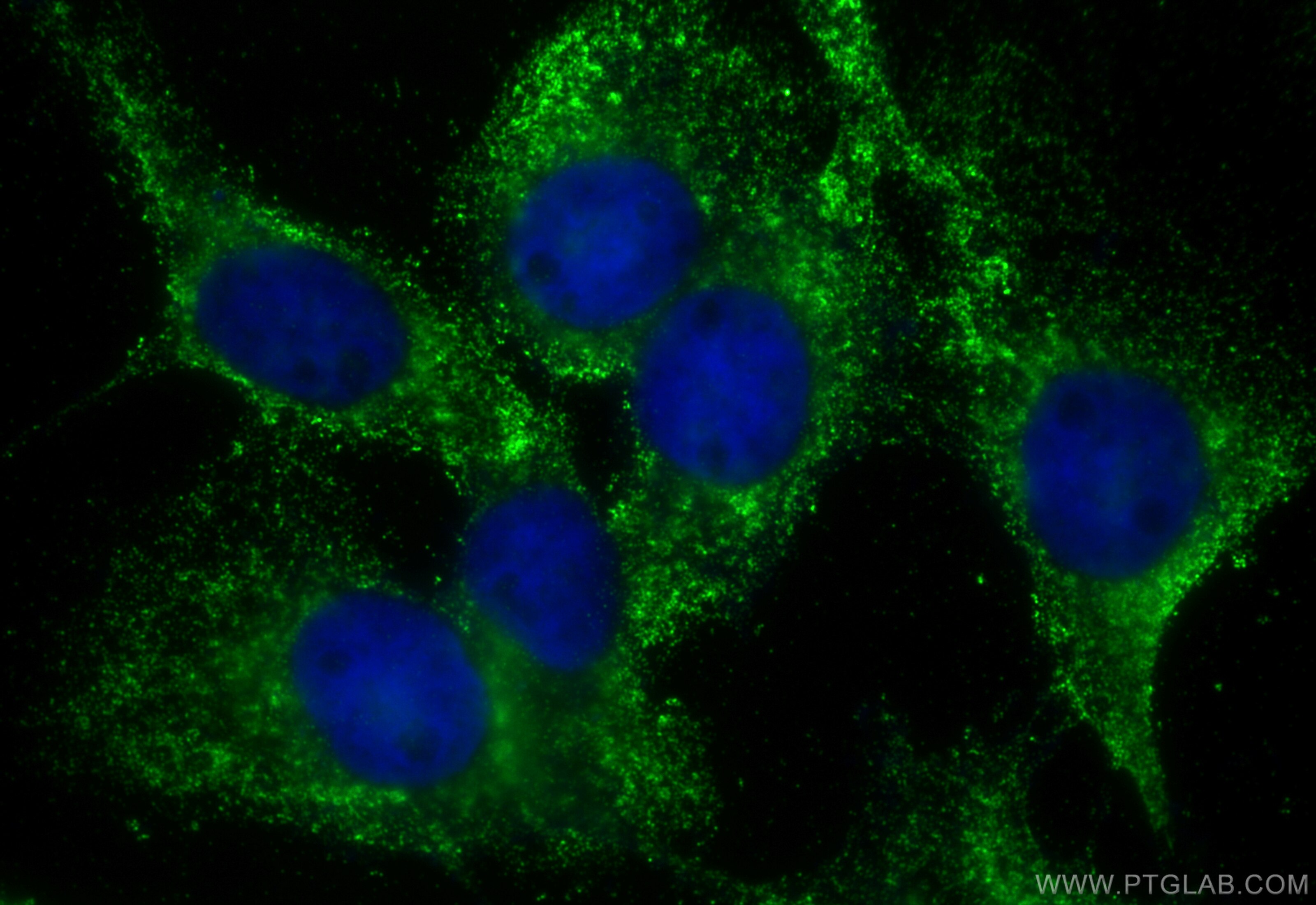
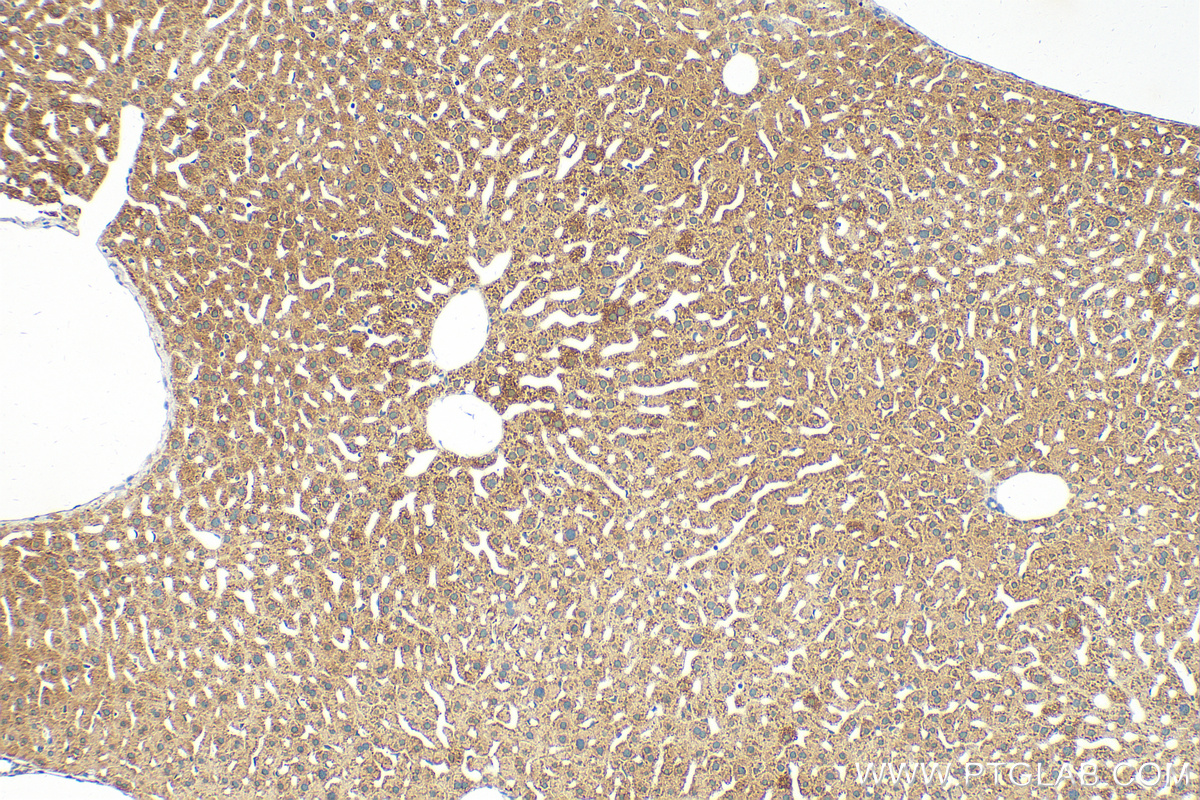
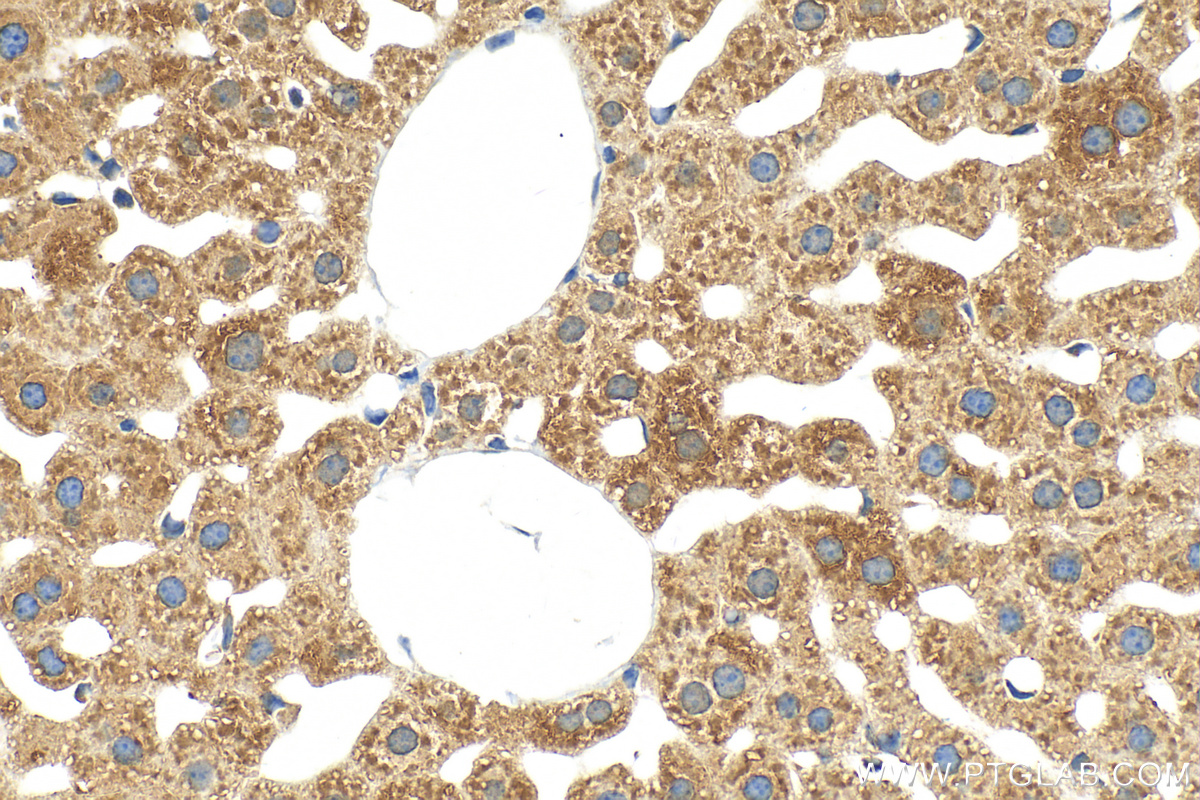
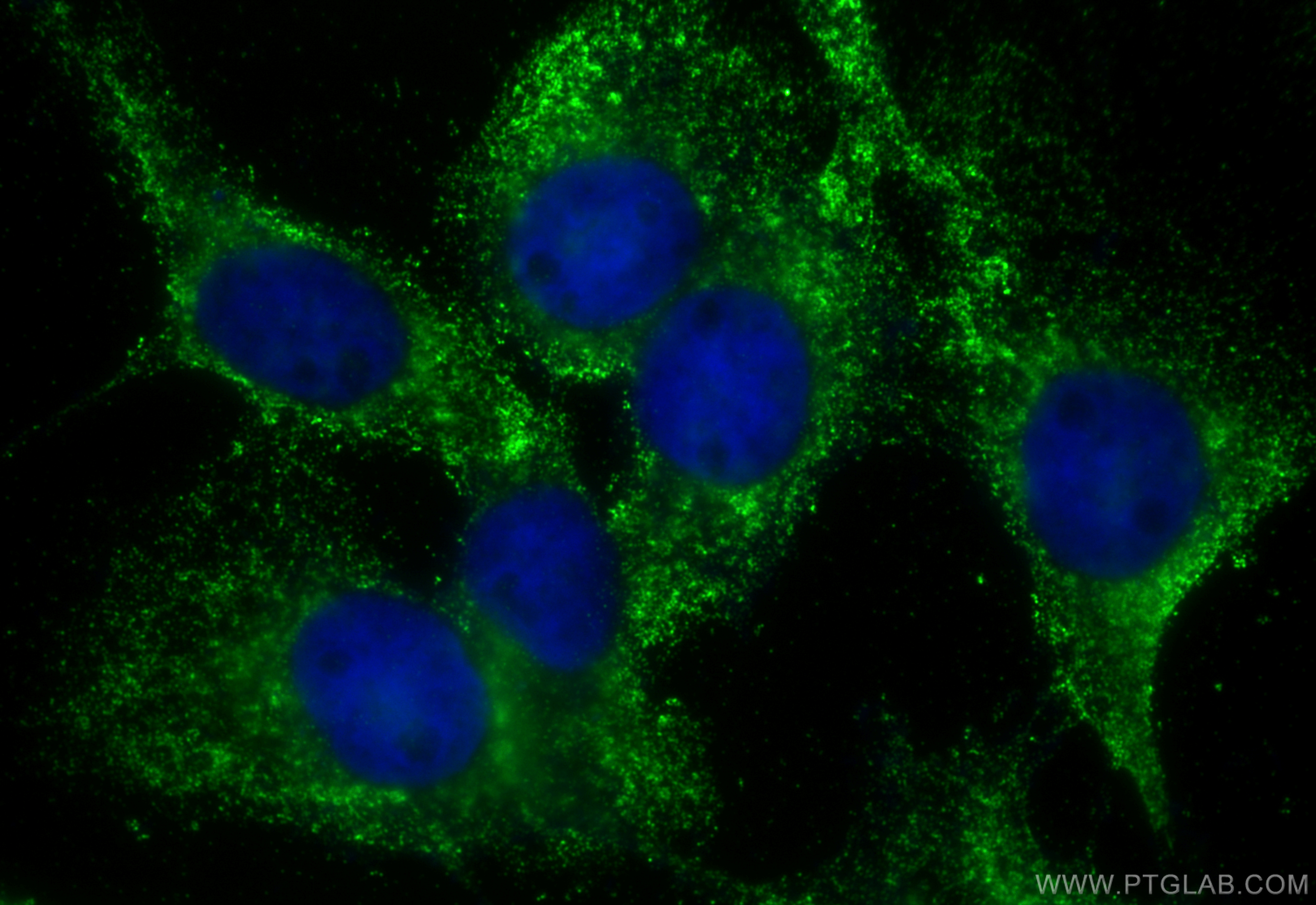
Tested Applications
| Positive IHC detected in | mouse liver tissue Note: suggested antigen retrieval with TE buffer pH 9.0; (*) Alternatively, antigen retrieval may be performed with citrate buffer pH 6.0 |
| Positive IF/ICC detected in | A431 cells |
Recommended dilution
| Application | Dilution |
|---|---|
| Immunohistochemistry (IHC) | IHC : 1:50-1:500 |
| Immunofluorescence (IF)/ICC | IF/ICC : 1:50-1:500 |
| It is recommended that this reagent should be titrated in each testing system to obtain optimal results. | |
| Sample-dependent, Check data in validation data gallery. | |
Product Information
31427-1-AP targets C20orf26 in IHC, IF/ICC, ELISA applications and shows reactivity with human, mouse samples.
| Tested Reactivity | human, mouse |
| Host / Isotype | Rabbit / IgG |
| Class | Polyclonal |
| Type | Antibody |
| Immunogen | C20orf26 fusion protein Ag35425 Predict reactive species |
| Full Name | chromosome 20 open reading frame 26 |
| GenBank Accession Number | BC031674 |
| Gene Symbol | C20orf26 |
| Gene ID (NCBI) | 26074 |
| RRID | AB_3669978 |
| Conjugate | Unconjugated |
| Form | Liquid |
| Purification Method | Antigen affinity Purification |
| UNIPROT ID | Q8NHU2 |
| Storage Buffer | PBS with 0.02% sodium azide and 50% glycerol , pH 7.3 |
| Storage Conditions | Store at -20°C. Stable for one year after shipment. Aliquoting is unnecessary for -20oC storage. 20ul sizes contain 0.1% BSA. |
Background Information
C20orf26, also known as CFAP61 (cilia and flagella associated protein 61), is a protein-coding gene located on human chromosome 20. The protein is predicted to be involved in cilium movement and cilium organization, and it is likely to be located in the axoneme and motile cilium. CFAP61 is also predicted to colocalize with the radial spoke stalk, which is a part of the structural framework of cilia and flagella.
Protocols
| Product Specific Protocols | |
|---|---|
| IHC protocol for C20orf26 antibody 31427-1-AP | Download protocol |
| IF protocol for C20orf26 antibody 31427-1-AP | Download protocol |
| Standard Protocols | |
|---|---|
| Click here to view our Standard Protocols |