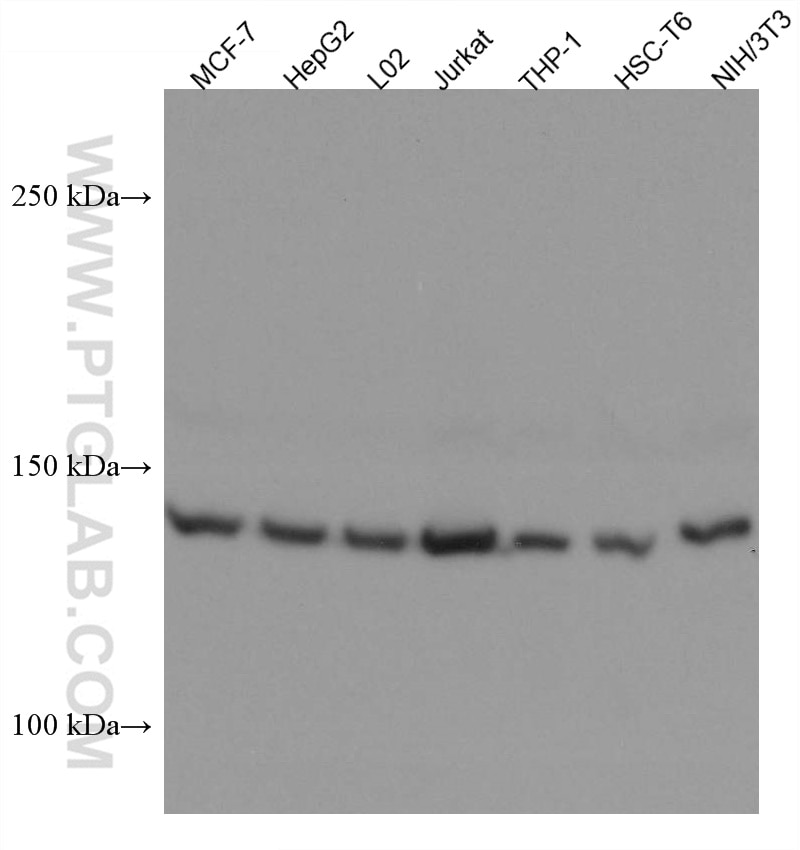
Tested Applications
| Positive WB detected in | MCF-7 cells, HepG2 cells, L02 cells, Jurkat cells, THP-1 cells, HSC-T6 cells, NIH/3T3 cells |
Recommended dilution
| Application | Dilution |
|---|---|
| Western Blot (WB) | WB : 1:5000-1:50000 |
| It is recommended that this reagent should be titrated in each testing system to obtain optimal results. | |
| Sample-dependent, Check data in validation data gallery. | |
Product Information
67436-1-Ig targets PEX1 in WB, ELISA applications and shows reactivity with human, mouse, rat samples.
| Tested Reactivity | human, mouse, rat |
| Host / Isotype | Mouse / IgG2b |
| Class | Monoclonal |
| Type | Antibody |
| Immunogen | PEX1 fusion protein Ag4623 Predict reactive species |
| Full Name | peroxisomal biogenesis factor 1 |
| Calculated Molecular Weight | 1283 aa, 143 kDa |
| Observed Molecular Weight | 143 kDa |
| GenBank Accession Number | BC035575 |
| Gene Symbol | PEX1 |
| Gene ID (NCBI) | 5189 |
| RRID | AB_2882673 |
| Conjugate | Unconjugated |
| Form | Liquid |
| Purification Method | Protein A purification |
| UNIPROT ID | O43933 |
| Storage Buffer | PBS with 0.02% sodium azide and 50% glycerol , pH 7.3 |
| Storage Conditions | Store at -20°C. Stable for one year after shipment. Aliquoting is unnecessary for -20oC storage. 20ul sizes contain 0.1% BSA. |
Background Information
Peroxin (PEX) proteins generate and maintain peroxisomes. The peroxin-1 (PEX1) ATPase facilitates the recycling of the peroxisome matrix protein receptor PEX5 and is the most commonly affected peroxin in human peroxisome biogenesis disorders (PMID: 28600347). PEX1 and PEX6 are the only members of the AAA family (for ATPases associated with diverse cellular) activities implicated in peroxisome biogenesis and are closely related to p97, which functions in endoplasmic reticulum-associated protein degradation to retrotranslocate endoplasmic reticulum proteins to the cytosol (PMID: 9695811; 11740563).
Protocols
| Product Specific Protocols | |
|---|---|
| WB protocol for PEX1 antibody 67436-1-Ig | Download protocol |
| Standard Protocols | |
|---|---|
| Click here to view our Standard Protocols |