The best anti-GFP antibody for immunoprecipitation: GFP-Trap
Comparison of GFP-Trap and anti-GFP IgG antibody
Life science laboratories apply green fluorescent proteins (GFP) to study protein localization, interaction and dynamics in fluorescence microscopy. Immunoprecipitation (IP), mass spectrometry (MS), co-immunoprecipitation (Co-IP) and/or affinity purification investigate more aspects including posttranslational modifications (PTMs), DNA binding, and protein-protein interaction. Here, we compare two different antibody systems for immunoprecipitation of GFP-fusion proteins: GFP-Trap and anti-GFP IgG antibody.
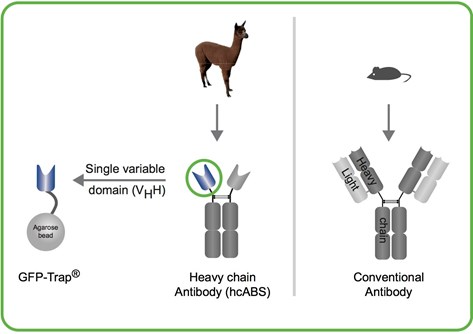
What are the structural differences between GFP-Trap and anti-GFP IgG antibody?
GFP-Trap consists of an anti-GFP VHH (green circle), derived from camelidae (alpaca, llama, camel, dromedaries, etc.) heavy chain antibodies. The anti-GFP VHH (also termed nanobody or single domain antibody) coupled to agarose beads is the GFP-Trap.
Anti-GFP IgG antibody is a commercially available mouse monoclonal antibody and has, like all IgGs, a large and complex structure:
Two heavy & two light chains.
Why is GFP-Trap the best antibody for immunoprecipitation?
- Clean – No contaminating heavy & light chains
- Pure – very low background due to reagent stability in harsh and stringent buffers
- Fast – favorable kinetic binding
- Reliable- strong binding due to its dissociation constant of 1 pM enables pulldowns of GFP-fusions expressed at low/endogenous levels
- Validated- Function and structure at highest level characterized
- Trust - Constant high quality – optimized recombinant production with care and thorough quality management
- Confidence – enormous amount of applications data available
- GFP-Trap sample – find out for yourself
Clean - NO heavy & light antibody chains in IP with GFP-Trap
The GFP-Trap provides clean pulldown results: You don’t have contaminations of heavy and light antibody chains, as you can see in figure below. GFP has been purified using both approaches—with the GFP-Trap (left) and an anti-GFP IgG antibody coupled to Protein A/G (right).
Input (I), non-bound (FT) and bound (B) fractions processed for IP were separated by SDS-PAGE followed by Coomassie staining and Western Blotting.
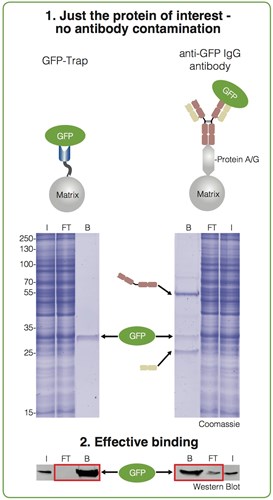
Immunoprecipitation of GFP from cell extracts. Precipitated GFP (green ellipse), denatured heavy (brown cartoon) and light chains (beige cartoon) of the IgG are marked by arrows.
The bound fraction of the GFP-Trap shows only the purified GFP. In contrast, the bound fraction of the conventional anti-GFP antibodies contains two additional strong bands besides the purified protein of interest. These bands at approximately 50 kDa and 25 kDa are contaminating heavy and light chains from the conventional anti-GFP antibody used for the immunoprecipitation. This may be a serious problem if you want to detect proteins of similar size.
Pure - Very low background due to reagent stability in harsh and stringent buffers
The following chemicals and temperatures can be applied to increase the stringency of binding and reduce background:
- 83°C
- 1mM DTT
- 2mM TCEP
- 3M Guanidinium•HCl
- 8M Urea
- 2M NaCl
- 2% Nonidet P40 Substitute
- 1% SDS
- 1% Triton X-100
Why is that?
The dissociation constant is the reciprocal measure of affinity. The high-affine, specific, and strong binding of our anti-GFP VHH to the GFP-fusion protein with an outstanding dissociation constant KD = 1 pM (1 picomolar or 1 ▪ 10-12 mol/l) allows to set binding conditions in a way that only protein of interest is recognized.
What applications need harsh binding conditions?
- Cleanest pulldowns
- Capture of endogenous level expressed proteins
- Detection of PTMs
- Reproducibility: e.g. for Mass Spec
- Stringent wash: e.g. ubiquitination assay see applications note here
- Alternative for streptavidin/biotin in Surface Plasmon Resonance
- Difficult binding, e.g. ChIP, RIP
A whitepaper discussing the chemical and thermal stability of the anti-GFP VHH: GFP-fusion protein complex can be found here.
Fast – GFP-Trap is ready to use and the binding of GFP-fusions occurs at a higher rate
GFP-Trap: As shown in the figure below, after addition of cell lysate to the GFP-Trap the immunoprecipitation takes place within a short incubation time. The entire workflow is typically completed within one hour because of high sensitivity, fast binding and extremely low dissociation constant. If performing Co-IP, less stable protein interaction partners are more likely co-precipitated due to shortened processing time.
anti-GFP IgG antibody: In contrast, when adding cell lysate to traditional antibodies against GFP, the incubation may take up to 12 hours or overnight. Plus, in a second step, the GFP antibody complex needs to be coupled to protein A/G beads, which takes another 1-2 hours. This makes the entire workflow to last typically more than 3 hours or (much) longer.
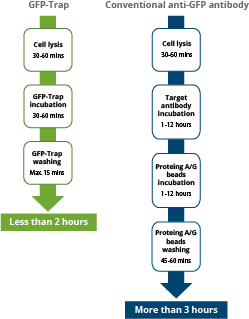
Workflow comparison between immunoprecipitations with alpaca Nanobody derived GFP-Trap (left) and with conventional anti-GFP IgG antibodies (right). The high specificity and affinity allow for considerable shorter incubation times.
Reliable & Reproducible - strong binding due to its dissociation constant of 1 pM enables pulldowns of GFP-fusions expressed at low/endogenous levels
Knowledge of the biophysical characteristics of an affinity matrix, i.e. its dissociation constant KD or its association/dissociation rates kon/koff, supports the design of an experiment.
The GFP-Trap has been intensively characterized and kinetic data are described in detail:
|
KD [M] |
kon [1/Ms] |
koff [s-1] |
|
1 ▪ 10-12 |
2.67 ▪ 107 |
3. 03 ▪ 10-5 |
Many proteins expressed at endogenous levels are at concentrations of 1-100 nM in a cell. Now, if the GFP-fusion protein of interest, e.g. inserted as a knock-in using GFP-tagging by CRISPR/Cas, is very difficult to express or expressed at sub-nanomolar levels, the GFP-Trap is by far the most efficient tool for immunoprecipitation.
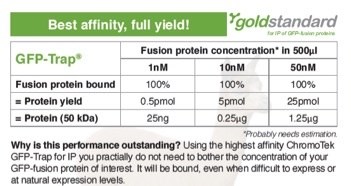
The amount of protein during immunoprecipitation (IP) depends on both protein concentration and dissociation constant KD. The GFP-Trap has a very high affinity and a phenomenal KD of 1 pM. Protein yields are stated for a total volume of 500 μl (sample, buffer & bead slurry). 5 pmol of a 50 kDa protein corresponds to 0.25 μg. This results in 500 μl to a concentration of 10 nM. Protein yields were calculated.
Anti-GFP IgG antibodies are not suited for IP of proteins expressed at low levels because their dissociation constants are orders of magnitude higher.
Validated - Function and structure at highest level characterized
The GFP-Trap is an anti-GFP VHH coupled to agarose beads. The anti-GFP VHH is a small, soluble and stable single polypeptide chain that is recombinantly expressed in bacteria virtually without lot-to-lot variations. This, in combination with stringent quality control procedures, makes its production robust and reproducible for reliable results.
The crystal structure of the GFP-Trap is available and its function in many applications has been tested. Protocols are available here.
- Immunoprecipitation (IP) and Co-IP
- Affinity purification
- Mass spectrometry
- Enzyme activity measurements
- RIP analysis
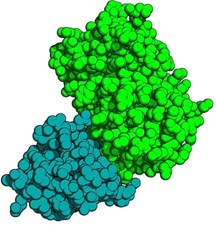
Crystal structure of anti-GFP VHH: Green Fluorescent Protein (GFP) complex. The anti-GFP-VHH is displayed blue, the GFP in green colors.
Specificity
ChromoTek tested GFP-Trap’s specificity with regards to fluorescent protein variants and derivatives.
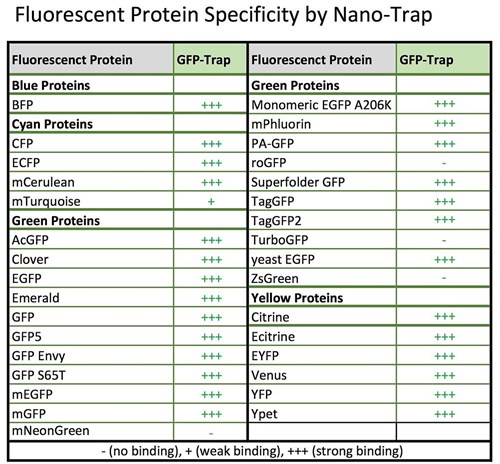
Specificity table GFP-Trap. An updated version of this table is here.
GFP-fusions from a multitude of cells and organisms have been analyzed using GFP-Trap to date. Here are some examples (in alphabetical order):
|
Aspergillus fumigatus |
Gallus domesticus |
Plasmodium berghei |
|
Arabidopsis thaliana |
Guinea pig cytomegalovirus (GPCMV) |
Petunia inflata |
|
Ashbya gossypii |
Human cell lines |
Plasmodium trophozoite |
|
Caenorhabditis elegans |
Leishmania |
Rattus norvegicus |
|
Danio rerio |
Magnaporthe oryzae |
Rice protoplast |
|
Dictyostelium discoideum |
Mammalian cell lines |
Sordaria macrospora |
|
Drosophila melanogaster |
Mus musculus |
Saccharomyces cerevisiae (yeast) |
|
E. coli |
Nicotiana benthamiana |
Tetrahymena thermophila |
|
Fusarium fujikuroi |
Oryza sativa |
Toxoplasma gondii |
For more organisms and details see ChromoTek’s GFP-Trap reference database.
Trust - Constant high quality – optimized production with care and thorough quality management
As mentioned above, The GFP-Trap anti-GFP VHH is recombinantly produced in E.coli. Every production lot is quality controlled and only product which fully matches specificity is released for sale and shipped to customers. Retention samples are taken on stock and are compared against any potentially failed product to identify potential errors in the process. A dedicated QC/QM laboratory and workforce has been established in early 2015.
Confidence – enormous amount of applications data available
There are more than 1,000 peer-reviewed articles referencing our GFP-Trap! The GFP-Trap is the most frequently cited monoclonal anti-GFP antibody, making it the gold standard for immunoprecipitation of GFP-fusion proteins.
See publications list on our website here or alternatively,see collection independently validated by citeab.com here.
Thanks to valuable feedback from our customers, we have developed new applications for the GFP-Trap. We are grateful to our customers in more than 60 countries for sharing their ideas and suggestions!
If you want to test the GFP-Trap yourself, request a free test sample here:
More information about GFP-Trap:
- ChromoTek Nanobodies
- How to plan an immunoprecipitation of your GFP-fusion protein
- GFP-Trap as affinity tag
- Fluorescent Protein Specificity for GFP-Trap and RFP-Trap
- Troubleshooting guide immunoprecipitation (IP)
- Literature References
- Elution strategies
- New limit in protein complex stability: GFP-binding protein:GFP complex
- On-bead digest protocol for mass spectrometry
- Ubiquitination of GFP-tagged proteins
- Capture Surface for Biacore assays
- Enzymatic activity assay
- GST- & GFP- nanobodies for bead-based protein arrays e.g. Luminex®
- How to prepare protein interaction partners for MS analysis
