SNAP-tag and CLIP-tag overview
SNAP-tag and CLIP-tag are self-labeling protein tags. The SNAP-tag protein and the CLIP-tag protein can be fused to a protein of interest (POI) and used for cellular and biochemical analysis.
- Which ligands for CLIP-tag are available?
- What are the differences between SNAP-tag, CLIP-tag and HaloTag?
- What are the differences between SNAP-tag, CLIP-tag and GFP?
- How can I detect my SNAP-tagged or CLIP-tagged protein in Western blot?
- How can I immunoprecipitate SNAP-tagged or CLIP-tagged proteins?
- Can I pulldown a SNAP-tag or CLIP-tag fusion proteins, that are already bound to a substrate/ ligand?
- How can I elute my SNAP-tag or CLIP-tag fusion protein?
- What are the binding kinetics of the SNAP/CLIP-tag-Trap?
- Excitation & Emission wavelengths of SNAP/CLIP-tag?
- Which ligands for CLIP-tag are available?
- What are the differences between SNAP-tag, CLIP-tag and HaloTag?
- What are the differences between SNAP-tag, CLIP-tag and GFP?
- How can I detect my SNAP-tagged or CLIP-tagged protein in Western blot?
- How can I immunoprecipitate SNAP-tagged or CLIP-tagged proteins?
- Can I pulldown a SNAP-tag or CLIP-tag fusion proteins, that are already bound to a substrate/ ligand?
- How can I elute my SNAP-tag or CLIP-tag fusion protein?
- What are the binding kinetics of the SNAP/CLIP-tag-Trap?
- Excitation & Emission wavelengths of SNAP/CLIP-tag?
What is SNAP-tag?
SNAP-tag is a self-labeling protein derived from human O6-alkylguanine-DNA-alkyltransferase. SNAP -Tag reacts with covalently with O6-benzylguanine derivatives, for example fluorescent dyes conjugated to guanine or chloropyrimidine. It can be used as a protein tag for tagging your protein of interest (POI).
What is CLIP-tag?
CLIP-tag is a modified version of SNAP-tag. It is also a self-labeling protein derived from human O6-alkylguanine-DNA-alkyltransferase. Instead of benzylguanine derivatives, CLIP tag is engineered to react with benzylcytosine derivatives. It can be used as a protein tag for tagging your protein of interest (POI). The following details refer to SNAP-tag but are also valid for CLIP. If there are differences between SNAP-tag and CLIP-tag they are listed.
How do SNAP-tag and CLIP-tag work?
After transfection with a plasmid encoding for the SNAP-tagged fusion protein, the SNAP-tagged protein is expressed in the cell line/ organism. Through incubation with specific ligands (see below point 9) your SNAP-tag fusion protein
-
-
turns fluorescent by binding a fluorescent dye or
-
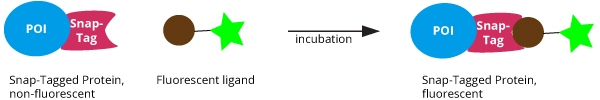
-
-
provides biotin for immobilization or other streptavidin modifications by binding a Biotin ligand.
-
Once the SNAP-tag has covalently bound the ligand, no other ligands can be bound as the catalytic center is blocked. No other application requiring a ligand can be performed. Alternatively, it is possible to immunoprecipitate or detect the SNAP tagged protein with antibodies or Nanobodies binding outside of the blocked catalytic center (see below point 13, 14, 15). For CLIP-tag, the same principle applies.
How is the complex between SNAP-tag and a ligand formed?
The SNAP-tag is derived from on human O6-alkylguanine-DNA-alkyltransferase (hATG), an enzyme involved in DNA repair. The SNAP-tag protein reacts with O6-benzylguanine (BG). Therefore ligands such as fluorescent dyes, biotin, etc., with guanine or a chloropyrimidine residue can serve as substrate for the SNAP-tag. During the labeling reaction, the substituted benzyl group of the ligand is covalently bound to the catalytic center of the SNAP-tag.
How is the complex between CLIP-tag and a ligand formed?
The CLIP-tag is derived from SNAP-tag. Instead of benzylguanine, CLIP-tag uses benzylcytosine as substrate. Therefore substrates such as fluorescent dyes, biotin, etc., with cytosin can be used with CLIP-tag. SNAP-tag and CLIP-tag can be simultaneously used in the same cell.
What sizes have SNAP-tag and CLIP-tag?
SNAP-tag consists of 182 amino acids (aa) and has a molecular weight of 19.4 kDa. CLIP-tag also consists of 182 aa and has a molecular weight of 19.4 kDa, too.
For what applications can SNAP-tag be used?
The SNAP-tag can be used for
-
- cell-based/imaging assays such as immunofluorescence
- immunoprecipitation
- protein purification.
For what applications can CLIP-tag be used?
The CLIP-tag can be used for
-
- cell-based/imaging assays such as immunofluorescence
- immunoprecipitation
Which ligands for SNAP-tag are available?
There are fluorescent dyes as substrates for microscopy available, which are cell permeable (TMR, Oregon Green) or cell impermeable such as Alexa Fluor 488, 546, and 647. In addition, there are Biotin and Maleimide as ligands available.
Which ligands for CLIP-tag are available?
As for SNAP-tag, there are cell permeable fluorescent dyes (TMR, CLIP-Cell™ 505) or cell impermeable fluorescent dyes such as CLIP-Surface™ 488, 547, and647. In addition, there is Biotin as substrate available.
What are the differences between SNAP-tag, CLIP-tag and HaloTag?
Halo-Tag is like SNAP-tag and CLIP-tag an extrinsically fluorescent protein. HaloTag is derived from the haloalkane dehalogenase DhaA from Rhodococcus rhodochrous and its active site reacts with chloroalkane linker substrates to form an irreversible bond.
Erdmann et al. (2019), compared SNAP-tag and HaloTag in live cell imaging with silicone rhodamine derivatives (=SiR-based dyes). They conclude that “for single-color imaging of SiR-based dyes , by either confocal or STED microscopy, we recommend Halo tagging as a first choice, since it provides fluorescence that is brighter and less prone to bleaching rapidly.”
|
Property |
SNAP-tag |
CLIP-tag |
HaloTag |
|
Discovery/ first publication |
2008 |
2008 |
2008 |
|
Origin |
human O6-alkylguanine-DNA-alkyltransferase |
human O6-alkylguanine-DNA-alkyltransferase |
Haloalkane dehalogenase from Rhodococcus rhodochrous |
|
Structure |
Monomer |
Monomer |
Monomer |
|
Reactivity |
O6-benzylguanine derivatives |
Benzylcytosine derivatives |
Chloroalkane derivatives |
|
Ligands |
Cell permeable or non-permeable dyes, fluorophores, biotin, and beads |
Cell permeable or non-permeable dyes, fluorophores, biotin, and beads |
Cell permeable or non-permeable dyes, fluorophores, biotin, and beads |
|
Excitation/ Emission maximum |
Depends on coupled fluorophore |
Depends on coupled fluorophore |
Depends on coupled fluorophore |
|
Length/ molecular weight (MW) |
182 amino acids, |
182 amino acids, |
297 amino acids, |
What are the differences between SNAP-tag, CLIP-tag and GFP?
GFP is an intrinsically fluorescent protein. The fluorescence arises through the protein itself, when excited with light (More technically speaking, GFP forms a β-barrel structure. The fluorescence comes from the backbone cyclization and oxidation of three amino acid residues in the middle of the barrel and results in a two-ring chromophore). There is no need for adding substrates or ligands in contrast to SNAP-tag and CLIP-tag. Photostability and quantum yield is normally lower compared to chemical dyes.
How can I detect my SNAP-tagged or CLIP-tagged protein in Western blot?
The sensitive anti-SNAP/CLIP-tag rat monoclonal antibody [6F9] from ChromoTek (now part of Proteintech) is validated for Western blotting.
How can I immunoprecipitate SNAP-tagged or CLIP-tagged proteins?
For efficient immunoprecipitation, our SNAP/CLIP-tag-Trap Agarose works very well. The SNAP/CLIP-tag-Trap consists of an anti-SNAP/CLIP-tag Nanobody conjugated to Agarose beads.
Can I pulldown a SNAP-tag or CLIP-tag fusion proteins, that are already bound to a substrate/ ligand?
Yes, you can pulldown a SNAP-tagged and CLIP-tagged proteins, even if the proteins are already bound to a ligand. The SNAP/CLIP-tag-Trap binds to epitope not overlapping with the catalytic center. Therefore, it is possible to conduct immunoprecipitation with SNAP-tagged and CLIP-tagged proteins that are already bound with the SNAP/CLIP-tag-Trap.
How can I elute my SNAP-tag or CLIP-tag fusion protein?
After pull-down with SNAP/CLIP-tag-Trap, one may elute the bound proteins using either 100 mM citric acid at pH3 or 200 mM glycine at pH 2.5.
What are the binding kinetics of the SNAP/CLIP-tag-Trap?
The binding kinetics of the anti-SNAP/CLIP-tag-Trap VHH have been characterized
Binding kinetics:
|
|
KD [M] |
kon(1/Ms) |
kdis(1/s) |
|
SNAP/CLIP-tag-Trap |
1.40.10-9 |
1.14.105 |
1.60.10-4 |
The SNAP/CLIP-tag-Trap allows for effective pull-down of low expression proteins.
Excitation & Emission wavelengths of SNAP/CLIP-tag?
There is no intrinsic fluorescence of the protein. Excitation and emission depend on the conjugated fluorescent dye. For example Alexa Fluor® 488 has an excitation maximum at 490 nm and an Emission maximum at 525 nm.
References
Labeling Strategies Matter for Super-Resolution Microscopy: A Comparison between HaloTags and SNAP-tags. (Erdmann et al., Cell Chemical Biology 2019) https://doi.org/10.1016/j.chembiol.2019.01.003
Related Content
Immunoprecipitation without additional band
How to conduct a Co-immunoprecipitation (Co-IP)
Which beads should I use for my immunoprecipitation?
Advantages and limitations of different antibody formats in immunoprecipitation
8 Top Tips For Immunoprecipitation
Learn how to save precious hours on your IP, IF, and western blotting experiments
Support
Request a Free Nano-Trap Sample
Nano-Trap system provides fast, reliable, and effective immunoprecipitation of fusion proteins. More than 3,000 peer-reviewed articles have already been published using ChromoTek's Nano-Traps.

