Affinity tags are very useful tools for protein purification. Fused to the protein of interest, they streamline the purification process by binding to a tag-specific resin. Obviously, tag selection is an important step as the purification tag can affect expression level, solubility, facilitate correct folding, protect from proteolysis, and re-direct proteins to a cellular compartment. In addition, the purification tag determines the affinity resin used.
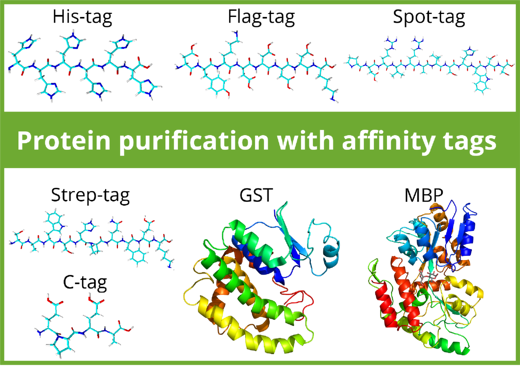
Purification tags can be split in two classes: Peptide tags and protein tags. Most peptide-tags are biochemically inert: they generally don’t affect protein folding and solubility and don’t interfere with protein function. In contrast, protein tags such as Maltose Binding Protein (MBP) and Glutathione-S-Transferase (GST) can increase the solubility of a protein and facilitate correct folding. Depending on the downstream application, protein tags may be removed by proteolytic cleavage after purification while peptide tags can remain at the protein of interest.
The use of affinity tags and resins influence the purity, number of purification steps, protein yield, elution efficiency, and the cost. Here is an overview over commonly used affinity tags in purification:
|
His-tag
- Consists of at least 6 histidine residues; often in combination with other purification tags
- Forms coordinated complexes (chelates) with central metal ions such as Ni2+ or Co2+
- Native elution with imidazole buffer
- Most commonly used affinity tag due to the high capacity of Ni- and Cobalt-resins combined with very low costs
- Rather high background level because Ni- or Co-resins also bind to endogenous proteins rich in histidines
- Can’t be used to purify metalloproteins
|
 |
|
FLAG®-tag
- Sequence: DYKDDDDK (8 amino acids) or
3X FLAG: DYKDHD-G-DYKDHD-I-DYKDDDDK with the final tag encoding an enterokinase cleavage site
- Artificially designed peptide-tag
- Binds to anti-FLAG antibody
- Native (competitive) elution with FLAG peptide or by low pH
- High costs: Relatively low binding capacity and low yield combined with limited regeneration of resin
|
 |
|
Spot-tag
- Sequence: PDRVRAVSHWSS (12 amino acids)
- Derived from beta-catenin, sequence optimized for binding to Spot-Tag Nanobody
- Binds to Spot-Tag Nanobody
- Native elution with low concentration of Spot peptide and pH shift
- High flexibility with regard to tag postion, buffer composition
- Cost effective: high binding capacity and possibility to re-use the resin
|
 |
|
C-Tag
- Sequence: EPEA (4 amino acids)
- C-terminal only
- Binds to anti-C-tag single-domain antibody fragment from camelids
- Native (competitive) elution with highly concentrated SEPEA peptide or high salt (e.g. 2 M MgCl2) and acidic solution
- Limited flexiblility due to C-terminal postion of tag
|
 |
|
Strep-tag®
- Sequence (Strep-tag II): WSHPQFEK (8 amino acids)
- Synthetic peptide-tag engineered for optimized binding to streptavidin
- Binds to streptavidin
- Native elution with desthiobiotin
- Poor binding of secreted Strap-tagged proteins due to biotin ingredients of cultivation media
- Sample contamination with biotinylated proteins from expression host
|
 |
|
GST-tag (Glutathione-S-Transferase)
- Size: 26 kDa (211 amino acids)
- GST is an abundant protein, involved in cellular defense
- Binds tightly to anti-GST Nanobody to lesser extend to glutathione resin
- Elution with reduced glutathione
- Increases solubility and expression of tagged protein
- Not suitable for multimeric protein complexes
- Large tag can interfere with target protein properties; therefore tag needs to be removed in additional purification step
|
 |
|
MBP-tag (Maltose binding protein)
- Size: 42 kDa (370 amino acids)
- Binds tightly to anti-MBP Nanobody to lesser extend to amylose resin
- Elution with maltose
- Increases solubility even better than GST-tag and facilitates correct protein folding
- Not suitable for multimeric protein complexes
- Large tag can interfere with target protein properties; therefore tag needs to be removed in additional purification step
|
 |







