Protein tags: Advantages and disadvantages
Understand when it is beneficial to tag a protein and when it is better to stay tag free.
Written by Sophie Williams, PhD researcher at the University of Bristol
The insertion of protein tags into recombinant proteins has become a useful and powerful tool in protein research. These tags can aid in the purification of the target protein from cell lysate, monitor the localization of the protein intracellularly, and aid in the detection of a previously uncharacterized protein. To date, there is a wide range of affinity tags to choose from, but can adding a non-native peptide or fusion protein tag have adverse effects?
Various protein tags are used routinely in research, including larger fusion-protein tags such as green fluorescent protein (GFP), maltose binding protein (MBP), and glutathione S-transferase (GST). Alternatively, smaller, more discrete peptides may be inserted into the recombinant target protein including a poly-histidine tag (His-tag) or a strep-tag. These tags are commonly added to either the N- or C-terminus of the target protein depending on the properties of the protein (e.g., the presence of subcellular localization sequences, downstream cleavage sites, or post-translational modification sites).
Applications of protein tags
When considering which protein tag to use in your research system, there are a few factors to consider:
-
Impact of the tag on the target protein’s structure and function
-
Requirement of a linker to separate the tag from the protein
-
Tag location
-
Inclusion of a protease cleavage site for tag removal
-
Final application of the protein
-
Required production level of recombinant protein
Along with this, some tags are more suited for specific purposes by incorporating desirable properties into your target protein.
Stability improvements
The production of tricky proteins, such as those requiring post-translational modifications, can be particularly troublesome in common expression hosts such as E. coli. As E. coli lacks the sophisticated machinery required for post-translational modifications found in many eukaryotic proteins, the expression of eukaryotic proteins in E. coli often results in protein misfolding and the production of inclusion bodies. As such, although up to 75% of human proteins can be expressed in E. coli, only 25% can be produced in the functional, soluble form1.
The addition of some protein tags may aid in the stability issues of troublesome proteins. For example, tags such as MBP, GST, and small ubiquitin related modifier (SUMO) are routinely fused with difficult proteins with poor solubility. These larger protein tags fold rapidly into stable proteins with good solubility after translation. The rapid folding of the protein tag can drive the folding of the target protein, aiding in the solubility and stability of the target protein while preventing its intracellular aggregation. This can be particularly important if high yields of recombinant protein are desired, such as during protein purification. For example, the human interferon IFN-γ was recently successfully expressed and purified in a functional form following SUMO-fusion, whereas previously it has aggregated into inclusion bodies in the native form2.
Protein-tag fusions can also aid in membrane protein production. Membrane proteins are traditionally complicated to express and purify, often becoming toxic to the expression host when overexpressed and requiring extraction from the cell membrane with detergents. They are also often unstable; however, it has been demonstrated that the fusion of protein tags may aid in membrane protein stability. For example, MBP has been fused to eukaryotic G-protein-coupled receptors to aid in expression and SUMO tags have been shown to aid in the folding and stability of both prokaryotic and eukaryotic membrane proteins3,4.
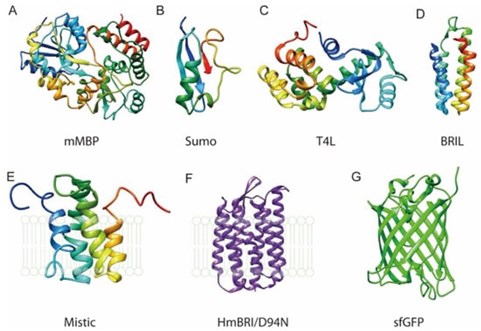
Figure 1. Examples of fusion protein tags to membrane protein purification. Figure taken from Liu and Li, Crystals (2022, open source)3.
Fluorescent signal
GFP and other related fluorescent protein tags provide a direct method to carry out subcellular localization studies relatively quickly and simply when applied with fluorescence imaging techniques. These fluorescent probes may be fused to the target protein to monitor its localization, abundance, and movements intracellularly with high spatial and temporal resolution.
Detection
GFP along with non-fluorescent tags such as FLAG® (DYKDDDDK), c-myc, or hemagglutinin antigen (HA) tags are widely used for protein detection during IP and western blotting studies. This can be particularly useful when working with an uncharacterized protein or a protein to which a good antibody has yet to be developed. Antibodies and Nanobodies to these tags are readily available and work excellently in western blotting, IP, and affinity purification.
Affinity
Fusion tags with high affinities to interaction partners such as small molecules (e.g., metal ions) or proteins (e.g., streptavidin, antibodies) may provide a useful method to purify the recombinant protein relatively simply and quickly. Affinity chromatography exploits this characteristic of numerous protein tags including the His-tag, Strep-tag, and GST. For example, the His-tag binds with high affinity to transition metal ions such as Ni2+ or Co2+, which can be immobilized on a solid support such as beads or resin. Once the His-tagged protein is bound to the metal-resin, contaminating proteins can be washed away leaving the desired protein that may be eluted in a high imidazole concentration buffer.
Drawbacks of inserting protein tags
Although the use of protein tags has revolutionized protein science, certain drawbacks need to be considered before tags are added:
1. Unknown impacts on the target protein
The insertion of protein tags (especially larger tags) may have unknown impacts on the structural and functional characteristics of the target protein. The insertion of a tag can lead to either partial or complete loss of function. For example, although small in tag-size, the insertion of a His-tag has resulted in significant reductions in activity in multiple enzymes in recent studies2,5. Therefore, to determine if the tag fusion is having an impact on the activity of the target protein, complementation studies to monitor the wild-type versus the recombinant protein’s activities would be required.
2. Protein instability upon tag removal
The removal of solubilizing fusion tags for downstream applications such as crystallization of the target protein may bring about the return of the target protein’s instability. As such, co-expression with a chaperone or antibody fragment binding may be a more appropriate approach if tag removal is required. Chaperones assist newly synthesized proteins in their correct folding while burying the target protein’s hydrophobic regions to prevent protein aggregation. As such, chaperones such as protein disulfide isomerase (PDI), GroES/L, or trigger factor (TF) are commonly co-expressed with troublesome proteins.
3. Extra steps of tag removal
Efficient removal of protein tags from the recombinant protein can often be difficult and time-consuming. Typically, this involves the use of a protease and the insertion of a protease cleavage site between the tag and protein, but the cleavage sequence may also influence the structure or function of the target protein.
4. Incorporation of non-native sequences
Some biotherapeutics require all protein tags to be removed (including linkers and protease cleavage sites) to leave only the native sequence. The presence of non-native sequences may interfere with the therapeutics function or may result in an immune response being raised when the drug is administered. As such, Proteintech’s Humankine® RUO and cGMP recombinant cytokines and growth factors are always tag-free.
Summary
Overall, each protein tag comes with its own benefits and hindrances. However, when the application and the target system are carefully considered, protein tags can provide an invaluable tool, saving researchers time when tackling tricky proteins.
|
|
Advantages |
Disadvantages |
Tagged Proteins |
· Ability to purify via tag’s affinity (e.g., affinity chromatography) · Improvements to solubility and stability of the target protein · Specific detection of protein tag, which may not be possible for tag-free protein · Increased total yield of protein through reduced degradation and increased expression |
·Tag may interfere with structure and biological activity of protein · Tag may need to be removed depending on downstream application · Instability of protein after tag removal · Larger tags impose a heavy metabolic burden on expression host |
Tag-free Proteins |
· Tag removal is not required · Native protein resulting in less modification to the structure and biological activity of the protein |
· Purification and detection are more challenging and complicated · Protein misfolding and insolubility will require additional steps to overcome, leading to lower yields |
Nanobodies against tags
Nano-Traps: affinity beads for immunoprecipitation
ChromoTek Nano-Traps are the benchmark in immunoprecipitation (IP) and allow fast and reliable one-step pulldowns of even low expressed proteins. They consist of Nanobodies coupled to beads and are ready-to-use. Low background, no extra bands & high specificity will improve your pulldown assay significantly.
Watch why GFP-Trap® gives the best results for immunoprecipitation:
Antibodies against tags
Highly cited antibodies against all popular tags. Browse here.
References:
- Costa, S., Almeida, A., Castro, A. & Domingues, L. Fusion tags for protein solubility, purification and immunogenicity in Escherichia coli: the novel Fh8 system. Frontiers in Microbiology vol. 5 at https://www.frontiersin.org/articles/10.3389/fmicb.2014.00063 (2014).
- Araújo, A. P. et al. Influence of the histidine tail on the structure and activity of recombinant chlorocatechol 1,2-dioxygenase. Biochem. Biophys. Res. Commun. 272, 480–484 (2000).
- Liu, S. & Li, W. Protein Fusion Strategies for Membrane Protein Stabilization and Crystal Structure Determination. Crystals vol. 12 at https://doi.org/10.3390/cryst12081041 (2022).
- Zuo, X. et al. Enhanced expression and purification of membrane proteins by SUMO fusion in Escherichia coli. J. Struct. Funct. Genomics 6, 103–111 (2005).
- Panek, A., Pietrow, O., Filipkowski, P. & Synowiecki, J. Effects of the polyhistidine tag on kinetics and other properties of trehalose synthase from Deinococcus geothermalis. Acta Biochim. Pol. 60, 163–166 (2013).
Related Content
Do you purify a special protein?
SNAP-tag and CLIP-tag overview
Immunoprecipitation of Myc-tagged proteins- How it works
Spot Capture and Detection Peptide Tag and Nanobody
Support
Newsletter Signup
Stay up-to-date with our latest news and events. New to Proteintech? Get 10% off your first order when you sign up.