Mitochondrial respiratory supercomplexes: a focus on COX7A isoforms
COX7A isoforms adapt the cellular energy production machinery to metabolic demands.
Written by Michele Brischigliaro, Ph.D. Postdoctoral Research Associate at the University of Miami Miller School of Medicine. Previously Dr Brischigliaro was a Postdoc for Dr Erika Fernandez-Vizarra, a leading mitochondrial researcher who recently presented her work (see recording here) on metabolic adaptions of OXPHOS organization and function to glycolytic switch using Proteintech’s antibodies.
What is the function of OXPHOS (oxidative metabolism)?
All living organisms transform energy to sustain life. Mitochondria are the organelles that provide most of the energy, in the form of adenosine triphosphate (ATP), in aerobic eukaryotic cells.
The term mitochondrion was originally coined in 1898 by German microbiologist Carl Benda. The word originates from the combination of two Greek words, mitos, meaning “thread,” and chondrion, meaning “grain,” as Benda described these organelles as rod-like structures with a tendency to form filaments. Despite the existence of different theories about the origin of mitochondria, convincing and widely accepted evidence supports the so-called endosymbiotic hypothesis. Accordingly, mitochondria derive from an ancestral organelle that originated from the stable integration of an endosymbiotic respiring α-proteobacterium1 into an anaerobic host cell related to Asgard archaea2. In eukaryotic cells, the energy contained in the nutrients is converted to metabolites that mitochondria utilize to produce ATP, the energetic “currency” that is needed for a plethora of cellular biochemical reactions.
The molecular machinery responsible for energy transformation is the oxidative phosphorylation (OXPHOS) system, which is composed of five multiprotein complexes embedded in the inner mitochondrial membrane. OXPHOS consists of two finely controlled processes: electron transport and ATP synthesis. Electron transport is based on a series of redox reactions that take place between complexes I–IV and two mobile electron carriers (coenzyme Q and cytochrome c). Electrons are taken by complexes I and II from reducing equivalents (NADH and FADH2, respectively). Electron transfer in complexes I, III, and IV is coupled with proton pumping from the mitochondrial matrix to the intermembrane space. Proton pumping generates an electrochemical gradient that provides the protonmotive force exploited by complex V to synthesize ATP.
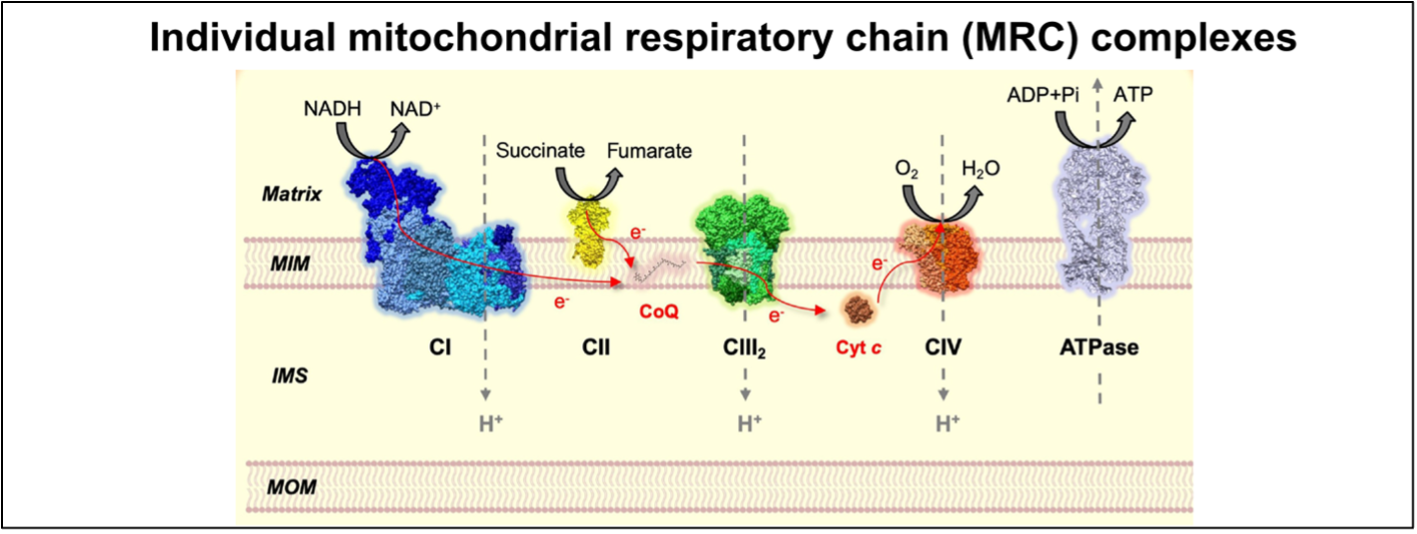
Figure 1. The mitochondrial respiratory chain (MRC) complexes and ATP synthase constitute the OXPHOS system. Figure adapted from Fernández-Vizarra & Ugalde, Trends in Biochemical Sciences (2022, Cell Press)16.
MOM = mitochondrial outer membrane, IMS = inner membrane space, MIM = mitochondrial inner membrane.
Importantly, the reducing equivalents in the form of NADH and FADH2 that feed the mitochondrial respiratory chain (MRC) are mostly generated through the Krebs cycle, aka the tricarboxylic acid (TCA) cycle, which is a central pathway where many metabolic routes converge. Thus, OXPHOS and the TCA cycle are well interconnected, and finely regulated mechanisms must exist for the mutual adaption of the two systems (Figure 2). A central metabolic route that derives metabolites from the TCA cycle is the conversion of pyruvate through the breakdown of glucose via glycolysis. Importantly, the rate-limiting enzyme that dictates the entry of pyruvate into the TCA cycle is the pyruvate dehydrogenase complex (PDH), which catalyzes the conversion of pyruvate to acetyl-coenzyme A (Acetyl-CoA) through pyruvate decarboxylation (Figure 2). The ability of pyruvate to enter the TCA cycle determines whether it will be used for oxidative metabolism or if, in turn, it will be converted into lactate through anaerobic fermentation.
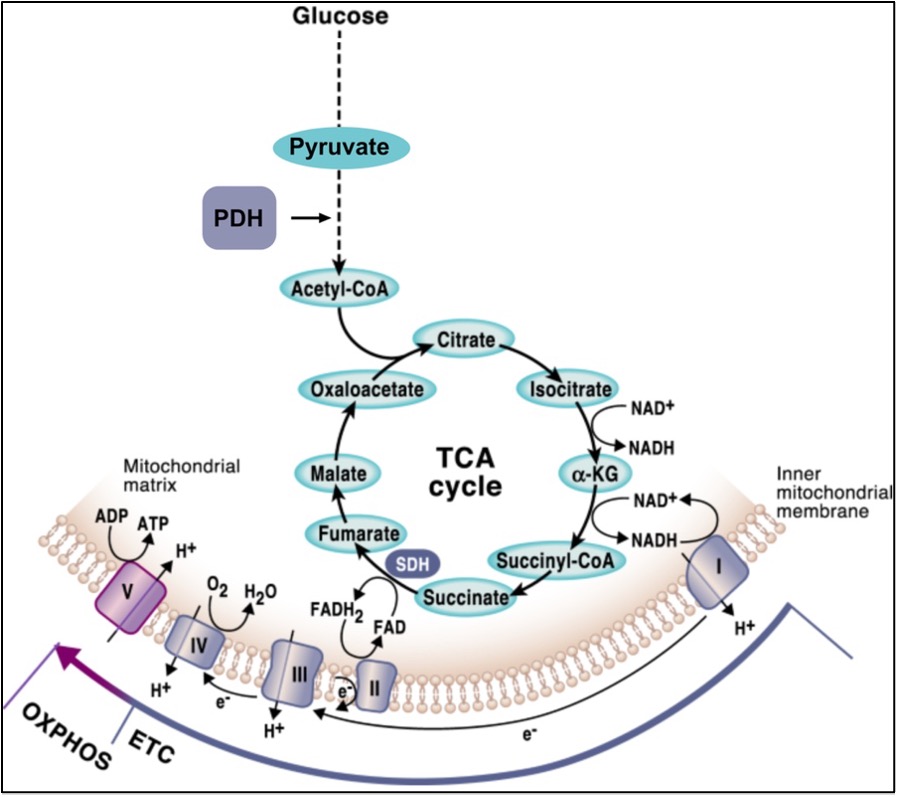
Figure 2. The mutual connection between OXPHOS and the TCA cycle. Figure from Martinez-Reyes & Chandel, Nature Communications (2020, open access).
Organization of the MRC: respiratory supercomplexes
The OXPHOS complexes exist as individual identities but can also associate to form different combinations of supramolecular structures known as “supercomplexes.” These OXPHOS entities were first described more than 20 years ago3 and their structure has recently been solved and described in different works4. Based on their molecular size and subunit composition, the main supercomplexes have been assigned the following stoichiometries: III2IV1, I1III2, and I1III2IV1, which is often referred to as the “respirasome” (Figure 3). In addition to these supercomplexes, complexes IV and V exist in dimeric forms.
One of the main determinants of supercomplex formation is the presence of different isoforms of a complex IV structural subunit, named COX7A. COX7A1 and COX7A2 are tissue-specific isoforms of the same subunit as they have different expression patterns depending on the cell type. COX7A1 is mainly expressed in muscle, whereas COX7A2 is ubiquitously expressed5. A third isoform exists in mammals: COX7A2L or COX7RP. This protein was renamed “SCAFI” (supercomplex-associated factor 1) because it was described as being mainly associated to supercomplexes6. SCAFI is a longer COX7A isoform that was initially thought to be essential for CIV binding to supercomplexes III2+IV and the respirasome7. The recent resolution of the mammalian supercomplexes III2+IV1 structure confirmed the structural role of SCAFI in the biogenesis of supercomplexes III2+IV18, providing a tether for CIV within the CIII2 structure, generating a strong interaction between the two complexes. However, the functional role of SCAFI in respirasome biogenesis is not central because the predominant respirasome species contains COX7A1/29. Notably, the loss of SCAFI does not severely affect the overall bioenergetic function of the MRC10,11, although it has been linked to certain metabolic alterations12,13,14.
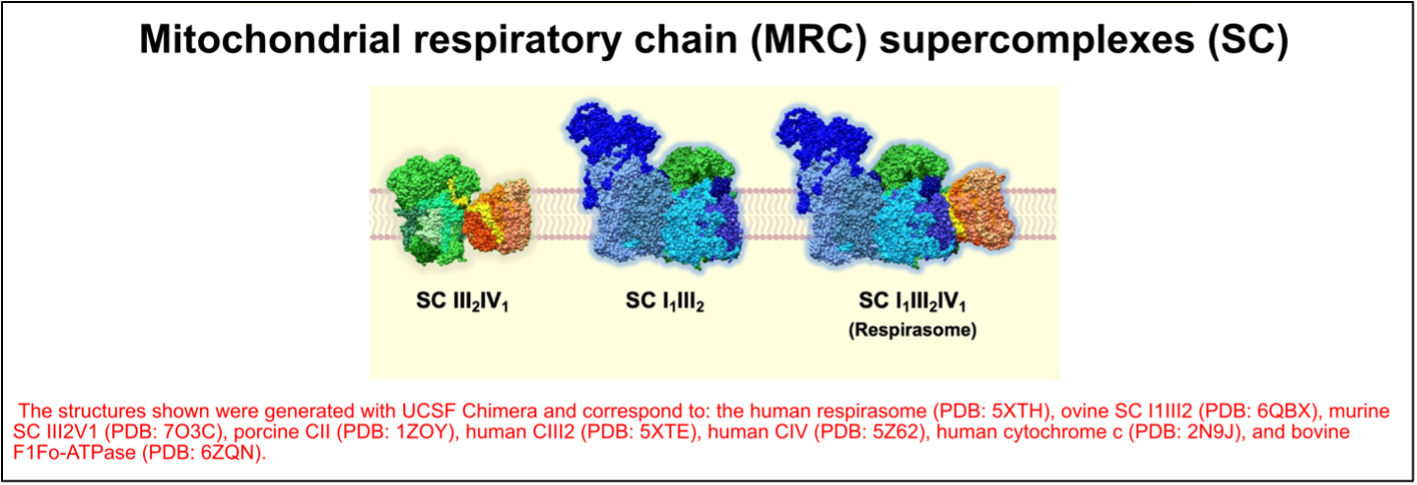
Figure 3. The organization of the mitochondrial respiratory chain (MRC) into supercomplexes (SC). Figure adapted from Fernández-Vizarra & Ugalde, Trends in Biochemical Sciences (2022, Cell Press)16.
Putative functional roles of the supercomplexes
Although the identity and structural interactions in the different supercomplex species are now well established, the functional reasons behind the existence of supercomplexes remain a matter of intense debate in the research field15,16. Different hypotheses concerning the role of supercomplexes have been proposed and are currently being tested.
The first possibility is that they are necessary for “substrate channeling,” i.e., their association allows the formation of enclosed pools of Q and cytochrome c, leading to increased electron transfer efficiency. However, the possibility of substrate channeling appears to be in contrast with kinetic and structural data. In fact, the available respirasome structures show that the distance between the catalytic sites of the enzymes is too high to allow substrate channeling8,17. The second main explanation to justify the existence of supercomplexes is that they play a structural function, stabilizing the complexes and/or serving as a platform for the efficient assembly of the individual complexes. The third and most recent hypothesis is that supercomplexes could be involved in the organizational adaption of the MRC, allowing changes in OXPHOS efficiency under different metabolic settings.
Metabolic regulation of MRC organization
The presence of SCAFI and COX7A1/2 is mutually exclusive, driving a different distribution of complex IV, as an individual entity and/or into different supercomplex species.
In cultured cells, COX7A2 is the main isoform, and when this isoform is missing SCAFI is overexpressed6. This leads to the re-organization of MRC with the specific formation of the so-called S-MRC organization, which includes SCAFI-containing complex IV, supercomplexes III2+IV1, and respirasome (S-respirasome). Importantly, this leads to less efficient OXPHOS6. Conversely, a lack of SCAFI leads to specific C-MRC formation, in which COX7A2 is part of complex IV and C-respirasome, and supercomplexes III2+IV1 do not form6. S-MRC and C-MRC coexist in physiological conditions, the latter being the predominant organization. In fact, the loss of SCAFI does not cause any functional defect of MRC. Importantly, C-MRC and S-MRC can be modulated in response to different metabolic conditions6. The C-respirasome and individual CIV are the preferred species under metabolic conditions in which the TCA is fully fueled by the enzymatic activity of the pyruvate dehydrogenase complex (PDH)6. Accordingly, cells lacking COX7A (expressing only SCAFI isoform) are unable to adapt their metabolism to the higher OXPHOS demands induced by the sustained activation of PDH6. Conversely, when the activity of the PDH is reduced (such as, for example, in cells from patients carrying mutations in PDH), metabolism is shifted towards glycolysis and the increase in SCAFI levels leads to the preferential formation of S-MRC6. In conclusion, the differential expression of the COX7A isoforms, i.e., COX7A2 and SCAFI, is a very important parameter to finely control MRC composition under different metabolic demands (Figure 4).
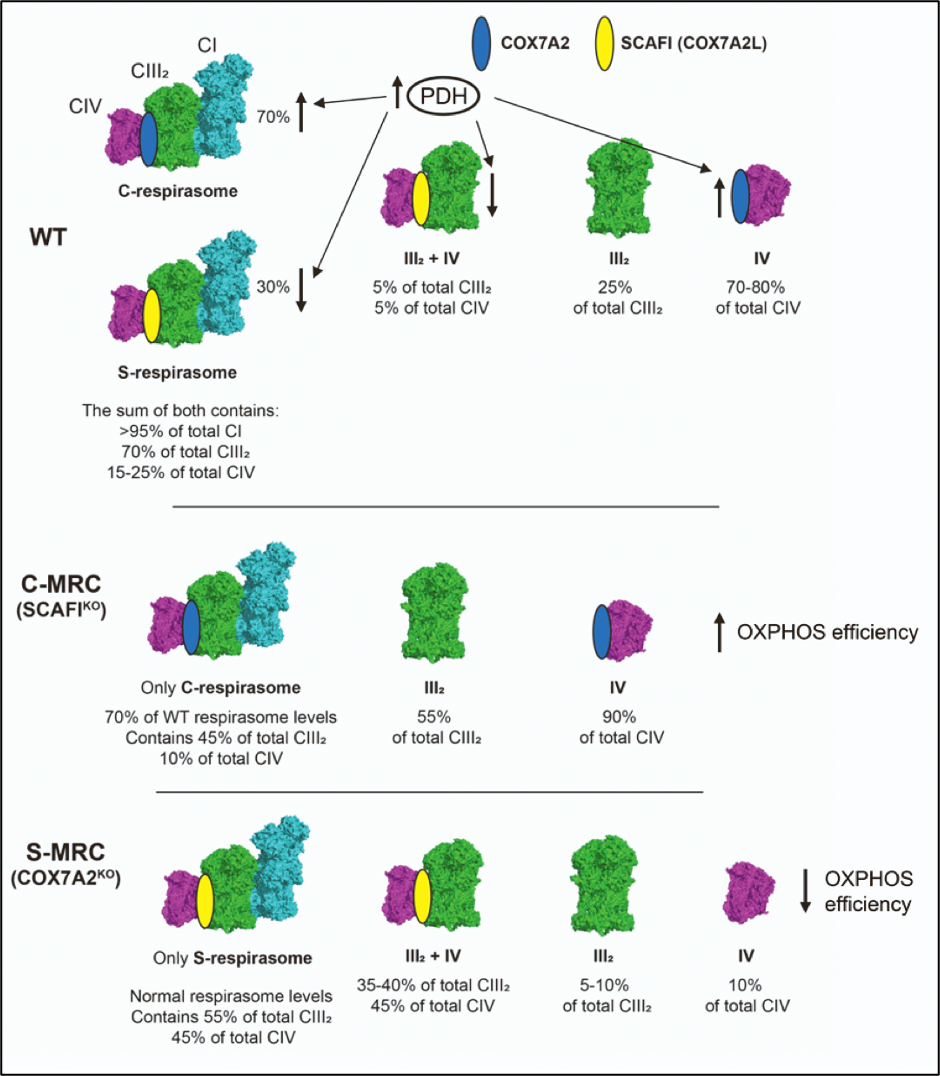
Figure 4. COX7A isoforms adapt the mitochondrial respiratory chain to metabolic switches. Figure adapted from Fernández-Vizarra et al., Cell Metabolism. (2022, Cell Press)6.
Antibodies for mitochondria research:
OXPHOS
|
Name |
Complex |
|
I |
|
|
II |
|
|
II |
|
|
IV |
|
|
V |
Popular mitochondria markers
COX7A isoforms
References
- Gray, M. W. Mosaic nature of the mitochondrial proteome: Implications for the origin and evolution of mitochondria. Proceedings of the National Academy of Sciences 112, 10133–10138 (2015).
- Asgard archaea illuminate the origin of eukaryotic cellular complexity | Nature. https://www.nature.com/articles/nature21031.
- Schägger, H. & Pfeiffer, K. Supercomplexes in the respiratory chains of yeast and mammalian mitochondria. The EMBO Journal 19, 1777–1783 (2000).
- Caruana, N. J. & Stroud, D. A. The road to the structure of the mitochondrial respiratory chain supercomplex. Biochem Soc Trans 48, 621–629 (2020).
- Sinkler, C. A. et al. Tissue- and Condition-Specific Isoforms of Mammalian Cytochrome c Oxidase Subunits: From Function to Human Disease. Oxidative Medicine and Cellular Longevity 2017, e1534056 (2017).
- Fernández-Vizarra, E. et al. Two independent respiratory chains adapt OXPHOS performance to glycolytic switch. Cell Metabolism 34, 1792-1808.e6 (2022).
- Lapuente-Brun, E. et al. Supercomplex Assembly Determines Electron Flux in the Mitochondrial Electron Transport Chain. Science 340, 1567–1570 (2013).
- Vercellino, I. & Sazanov, L. A. Structure and assembly of the mammalian mitochondrial supercomplex CIII2CIV. Nature 598, 364–367 (2021).
- Fernández-Vizarra, E. et al. SILAC-based complexome profiling dissects the structural organization of the human respiratory supercomplexes in SCAFIKO cells. Biochim Biophys Acta Bioenerg 1862, 148414 (2021).
- Mourier, A., Matic, S., Ruzzenente, B., Larsson, N.-G. & Milenkovic, D. The respiratory chain supercomplex organization is independent of COX7a2l isoforms. Cell Metab 20, 1069–1075 (2014).
- Lobo-Jarne, T. et al. Human COX7A2L Regulates Complex III Biogenesis and Promotes Supercomplex Organization Remodeling without Affecting Mitochondrial Bioenergetics. Cell Reports 25, 1786-1799.e4 (2018).
- Calvo, E. et al. Functional role of respiratory supercomplexes in mice: SCAF1 relevance and segmentation of the Qpool. Sci Adv 6, eaba7509 (2020).
- García-Poyatos, C. et al. Scaf1 promotes respiratory supercomplexes and metabolic efficiency in zebrafish. EMBO Rep 21, e50287 (2020).
- Berndtsson, J. et al. Respiratory supercomplexes enhance electron transport by decreasing cytochrome c diffusion distance. EMBO reports 21, e51015 (2020).
- Lobo-Jarne, T. & Ugalde, C. Respiratory Chain Supercomplexes: Structures, Function and Biogenesis. Semin Cell Dev Biol 76, 179–190 (2018).
- Fernández-Vizarra, E. & Ugalde, C. Cooperative assembly of the mitochondrial respiratory chain. Trends in Biochemical Sciences 47, 999–1008 (2022).
- Hirst, J. Open questions: respiratory chain supercomplexes—why are they there and what do they do? BMC Biology 16, 111 (2018).
Related Content
Metabolic Adaptations of OXPHOS Organization and Function to Glycolytic Switch
Featured Mitochondria Antibodies
Antibodies for metabolism research
Brown adipose tissue product feature
Detecting low abundance proteins via western blot
Support
Newsletter Signup
Stay up-to-date with our latest news and events. New to Proteintech? Get 10% off your first order when you sign up.