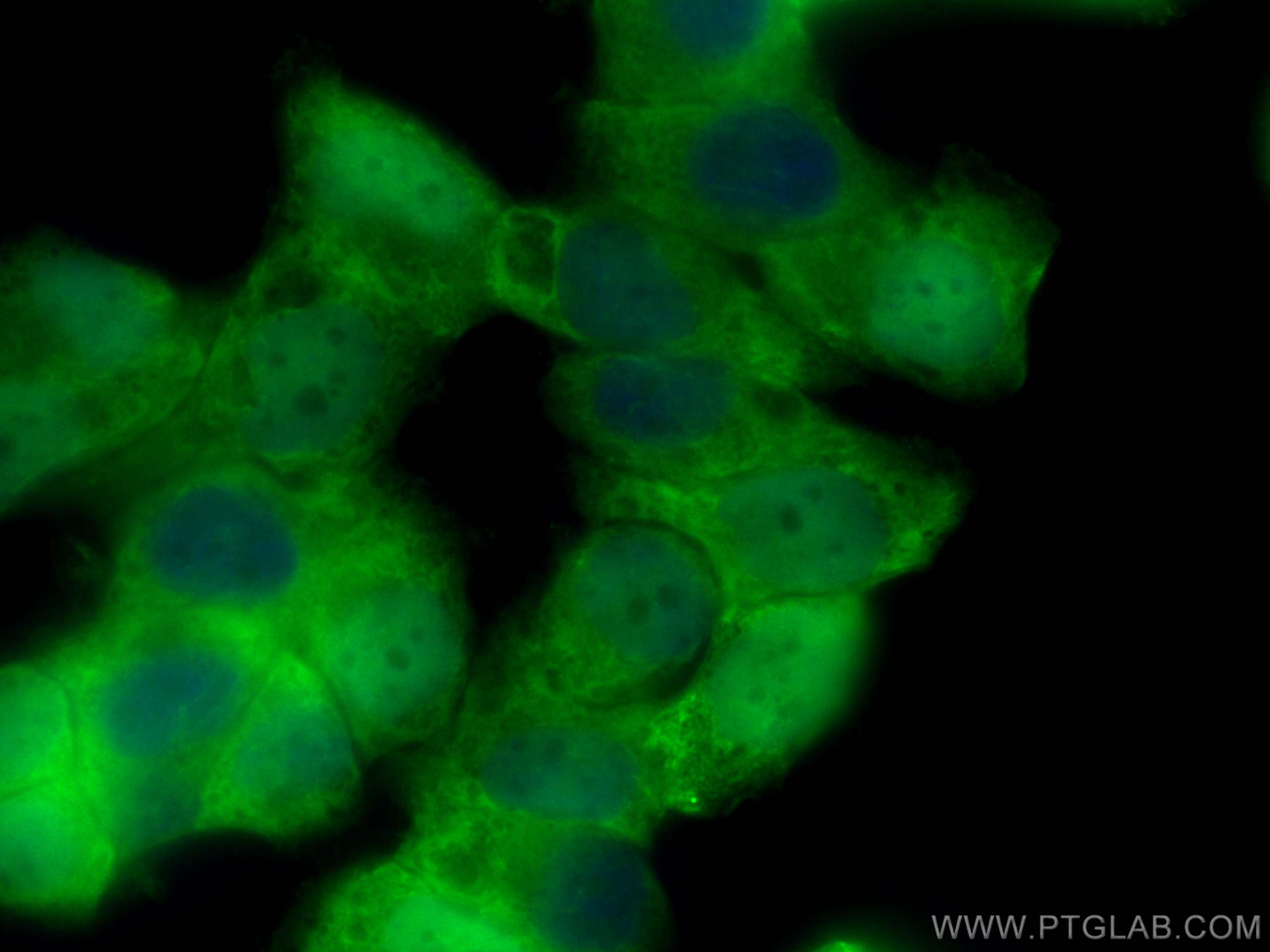
Tested Applications
| Positive IF/ICC detected in | NCCIT cells |
Recommended dilution
| Application | Dilution |
|---|---|
| Immunofluorescence (IF)/ICC | IF/ICC : 1:50-1:500 |
| It is recommended that this reagent should be titrated in each testing system to obtain optimal results. | |
| Sample-dependent, Check data in validation data gallery. | |
Product Information
CL488-68071 targets STRA8 in IF/ICC applications and shows reactivity with Human, rabbit, mouse samples.
| Tested Reactivity | Human, rabbit, mouse |
| Host / Isotype | Mouse / IgG1 |
| Class | Monoclonal |
| Type | Antibody |
| Immunogen | STRA8 fusion protein Ag29680 Predict reactive species |
| Full Name | stimulated by retinoic acid gene 8 homolog (mouse) |
| Calculated Molecular Weight | 37 kDa |
| Observed Molecular Weight | 43-60 kda |
| GenBank Accession Number | NM_182489 |
| Gene Symbol | STRA8 |
| Gene ID (NCBI) | 346673 |
| RRID | AB_3084394 |
| Conjugate | CoraLite® Plus 488 Fluorescent Dye |
| Excitation/Emission Maxima Wavelengths | 493 nm / 522 nm |
| Form | Liquid |
| Purification Method | Protein G purification |
| UNIPROT ID | Q7Z7C7 |
| Storage Buffer | PBS with 50% Glycerol, 0.05% Proclin300, 0.5% BSA, pH 7.3. |
| Storage Conditions | Store at -20°C. Avoid exposure to light. Stable for one year after shipment. Aliquoting is unnecessary for -20oC storage. |
Background Information
Stimulated by retinoic acid 8 (Stra8), one of genes induced by retinoic acid (RA), is required for the meiotic initiation of male spermatogenesis. Stimulated by Retinoic Acid Gene 8 (STRA8) protein has been proposed as the molecular effector of Retinoic Acid (RA) involved in meiosis-inductive activity. Stra8 is a 45-kDa vertebrate-specific gene, without significant homologous proteins, expressed in the cytoplasm and nuclei of germ cells between mitosis and meiosis and presents exclusively in the embryonic and postnatal gonads of mammals. (PMID: 30106128, PMID: 31253359)
Protocols
| Product Specific Protocols | |
|---|---|
| IF protocol for CL Plus 488 STRA8 antibody CL488-68071 | Download protocol |
| Standard Protocols | |
|---|---|
| Click here to view our Standard Protocols |