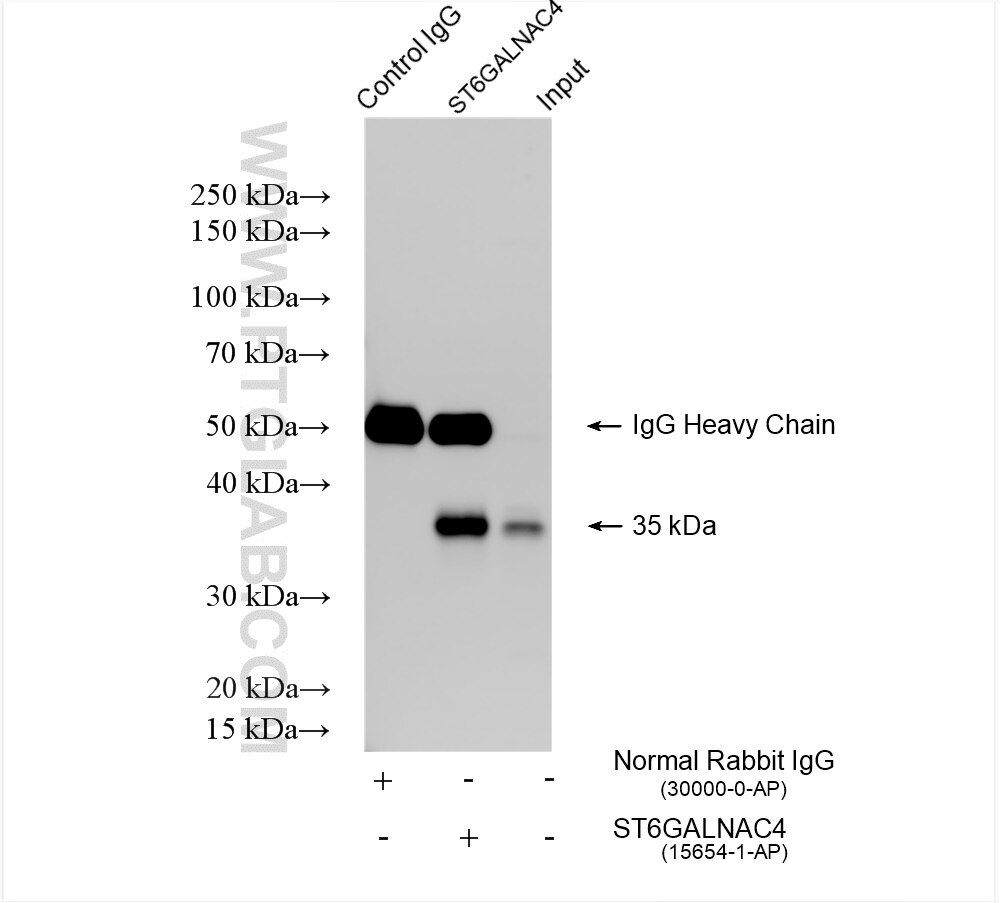
Tested Applications
| Positive IP detected in | HeLa cells |
Recommended dilution
| Application | Dilution |
|---|---|
| Immunoprecipitation (IP) | IP : 0.5-4.0 ug for 1.0-3.0 mg of total protein lysate |
| It is recommended that this reagent should be titrated in each testing system to obtain optimal results. | |
| Sample-dependent, Check data in validation data gallery. | |
Product Information
15654-1-AP targets ST6GALNAC4 in IP, ELISA applications and shows reactivity with human samples.
| Tested Reactivity | human |
| Host / Isotype | Rabbit / IgG |
| Class | Polyclonal |
| Type | Antibody |
| Immunogen | ST6GALNAC4 fusion protein Ag7995 Predict reactive species |
| Full Name | ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 4 |
| Calculated Molecular Weight | 34 kDa |
| Observed Molecular Weight | 35 kDa |
| GenBank Accession Number | BC036705 |
| Gene Symbol | ST6GALNAC4 |
| Gene ID (NCBI) | 27090 |
| RRID | AB_3669218 |
| Conjugate | Unconjugated |
| Form | Liquid |
| Purification Method | Antigen affinity purification |
| UNIPROT ID | Q9H4F1 |
| Storage Buffer | PBS with 0.02% sodium azide and 50% glycerol , pH 7.3 |
| Storage Conditions | Store at -20°C. Stable for one year after shipment. Aliquoting is unnecessary for -20oC storage. 20ul sizes contain 0.1% BSA. |
Background Information
Alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3-N-acetyl-galactosaminide alpha-2,6-sialyltransferase (also known as ST6GALNAC4) is a member of the sialyltransferases, which catalyzes the transfer of sialic acid from cytidine monosphosphate (CMP)-sialic acid to galactose-containing substrates. ST6GALNAC4 may be induced abnormal TGFBR2 glycosylation, resulting in the higher protein levels of TGFBR2 and TGF pathway increased activation (PMID: 37381011, 37034303). ST6GALNAC4 was reported to influence patient prognosis though subverting immunosurveillance in Chronic lymphocytic leukemia.
Protocols
| Product Specific Protocols | |
|---|---|
| IP protocol for ST6GALNAC4 antibody 15654-1-AP | Download protocol |
| Standard Protocols | |
|---|---|
| Click here to view our Standard Protocols |