Tested Applications
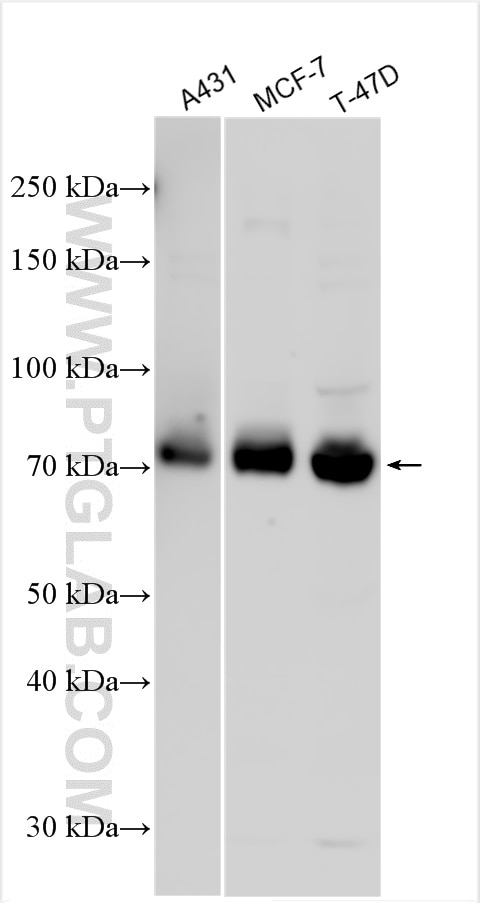
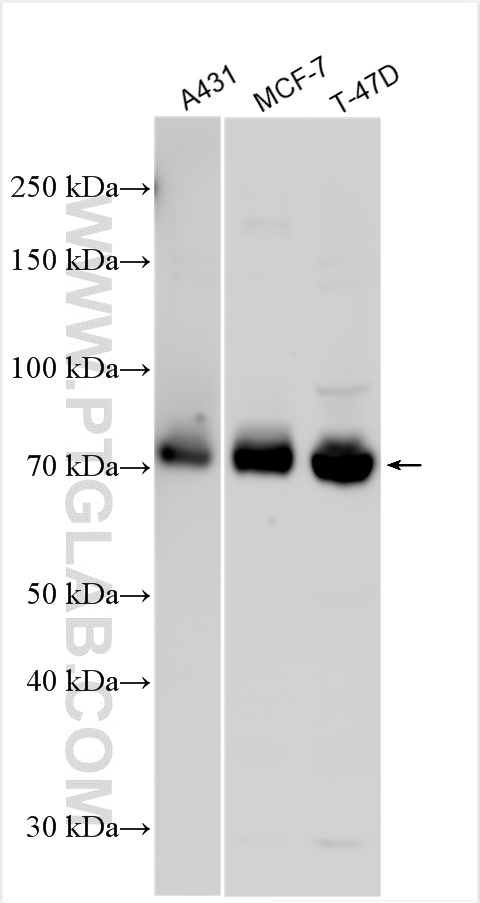
| Positive WB detected in | A431 cells, MCF-7 cells, T-47D cells |
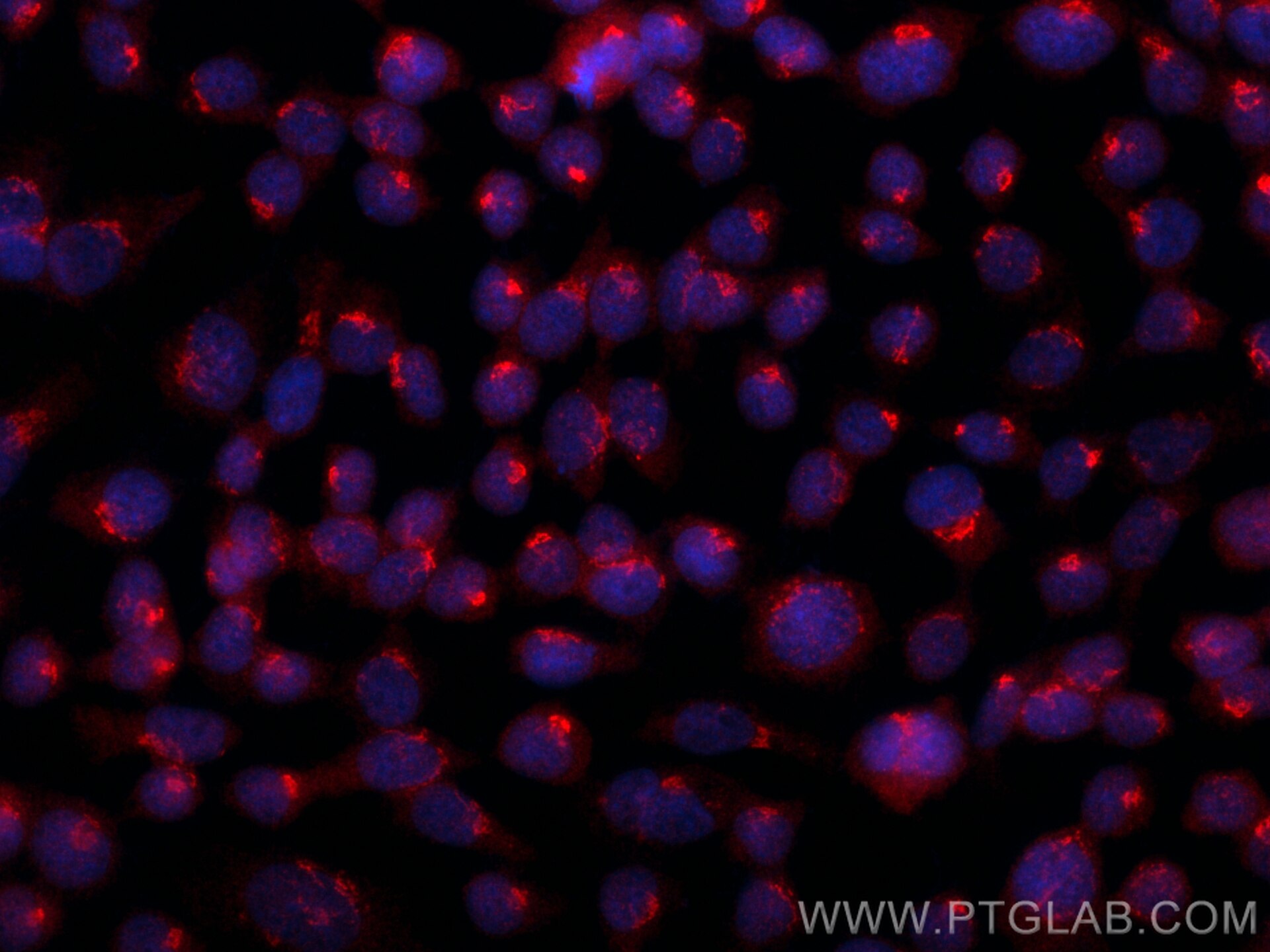
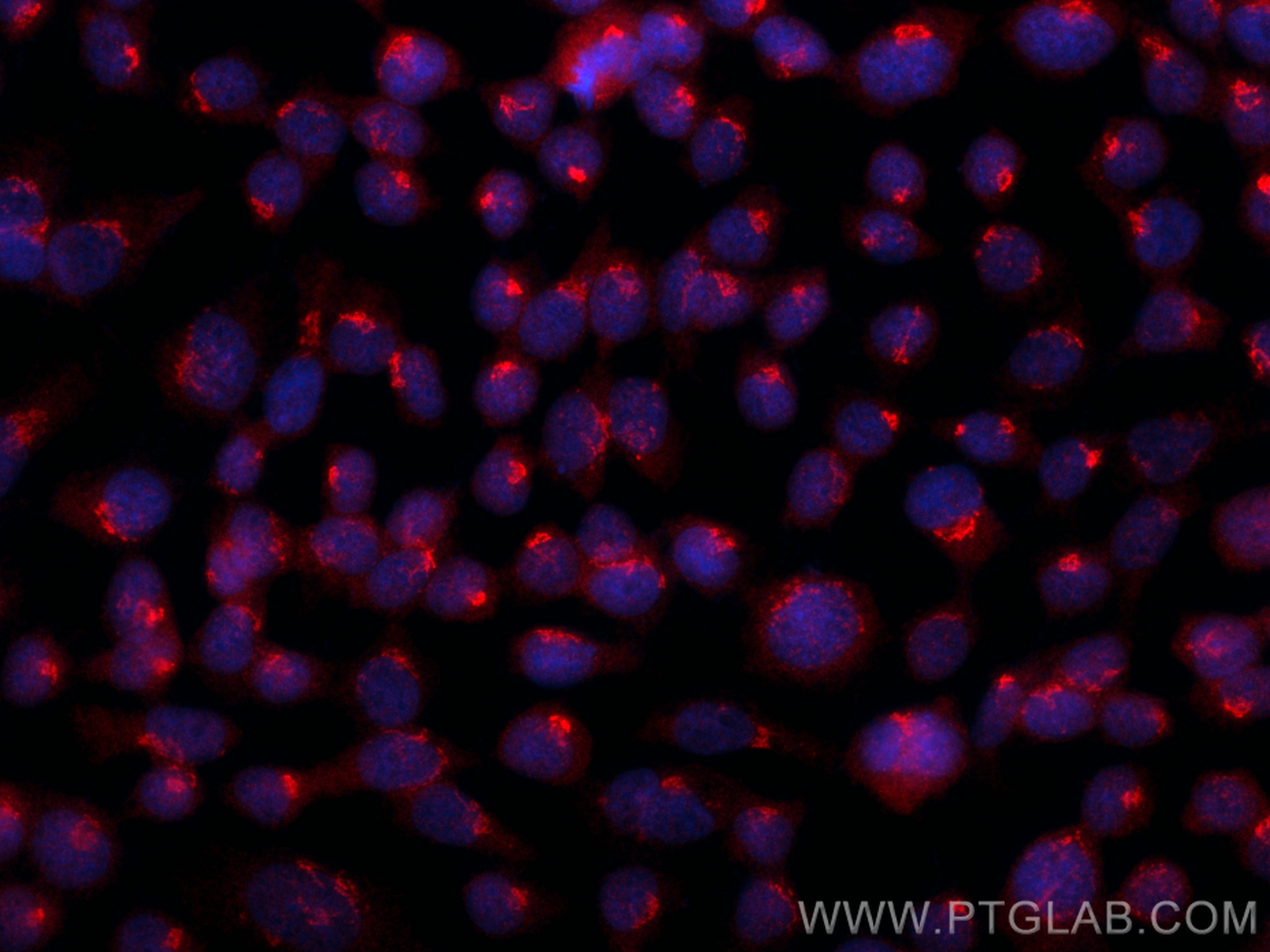
| Positive IF/ICC detected in | A431 cells |
Recommended dilution
| Application | Dilution |
|---|---|
| Western Blot (WB) | WB : 1:2000-1:12000 |
| Immunofluorescence (IF)/ICC | IF/ICC : 1:200-1:800 |
| It is recommended that this reagent should be titrated in each testing system to obtain optimal results. | |
| Sample-dependent, Check data in validation data gallery. | |
Product Information
13671-1-AP targets GALNT6 in WB, IF/ICC, ELISA applications and shows reactivity with human samples.
| Tested Reactivity | human |
| Host / Isotype | Rabbit / IgG |
| Class | Polyclonal |
| Type | Antibody |
| Immunogen | GALNT6 fusion protein Ag4652 Predict reactive species |
| Full Name | UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 6 (GalNAc-T6) |
| Calculated Molecular Weight | 622 aa, 71 kDa |
| Observed Molecular Weight | 71 kDa |
| GenBank Accession Number | BC035822 |
| Gene Symbol | GALNT6 |
| Gene ID (NCBI) | 11226 |
| RRID | AB_3669168 |
| Conjugate | Unconjugated |
| Form | Liquid |
| Purification Method | Antigen affinity purification |
| UNIPROT ID | Q8NCL4 |
| Storage Buffer | PBS with 0.02% sodium azide and 50% glycerol , pH 7.3 |
| Storage Conditions | Store at -20°C. Stable for one year after shipment. Aliquoting is unnecessary for -20oC storage. 20ul sizes contain 0.1% BSA. |
Background Information
GALNT6 (polypeptide N-acetylgalactosaminyltransferase 6, GalNAc-T6), which is involved in the initiation of O-glycosylation, has been reported to play crucial roles in mammary carcinogenesis through binding to several substrates (PMID: 32393740). GALNT6 is upregulated in breast, ovarian, renal cell, lung and pancreatic cancers (PMID: 20215525). Overexpression of GRP78 could enhance Golgi-to-ER relocation of GALNT6 (PMID: 28110670).
Protocols
| Product Specific Protocols | |
|---|---|
| WB protocol for GALNT6 antibody 13671-1-AP | Download protocol |
| IF protocol for GALNT6 antibody 13671-1-AP | Download protocol |
| Standard Protocols | |
|---|---|
| Click here to view our Standard Protocols |