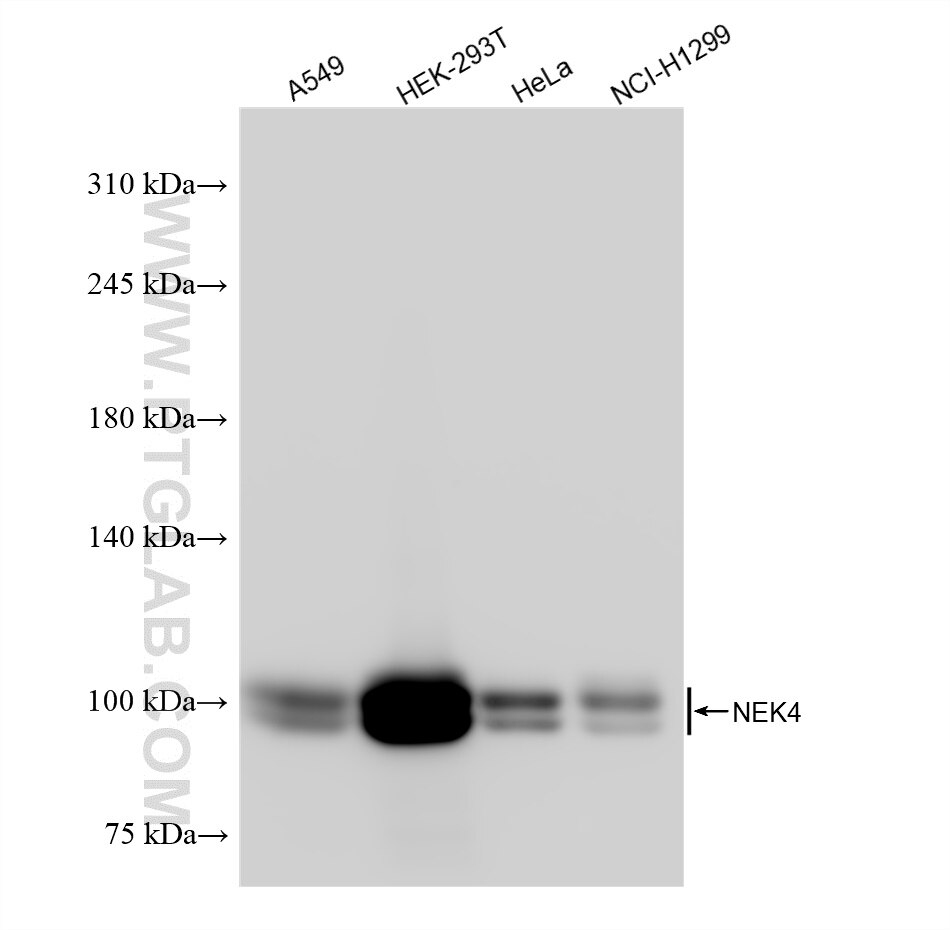
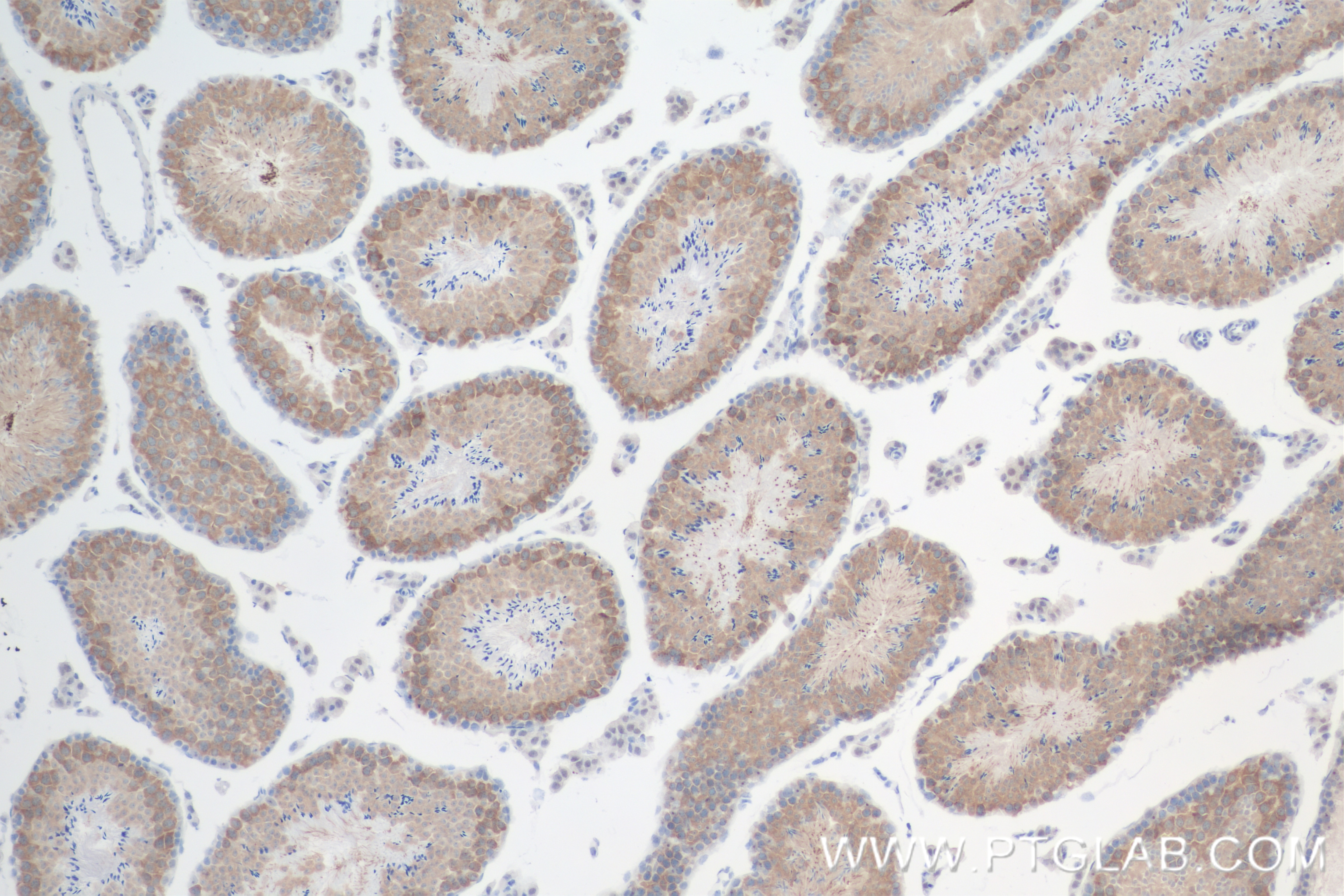
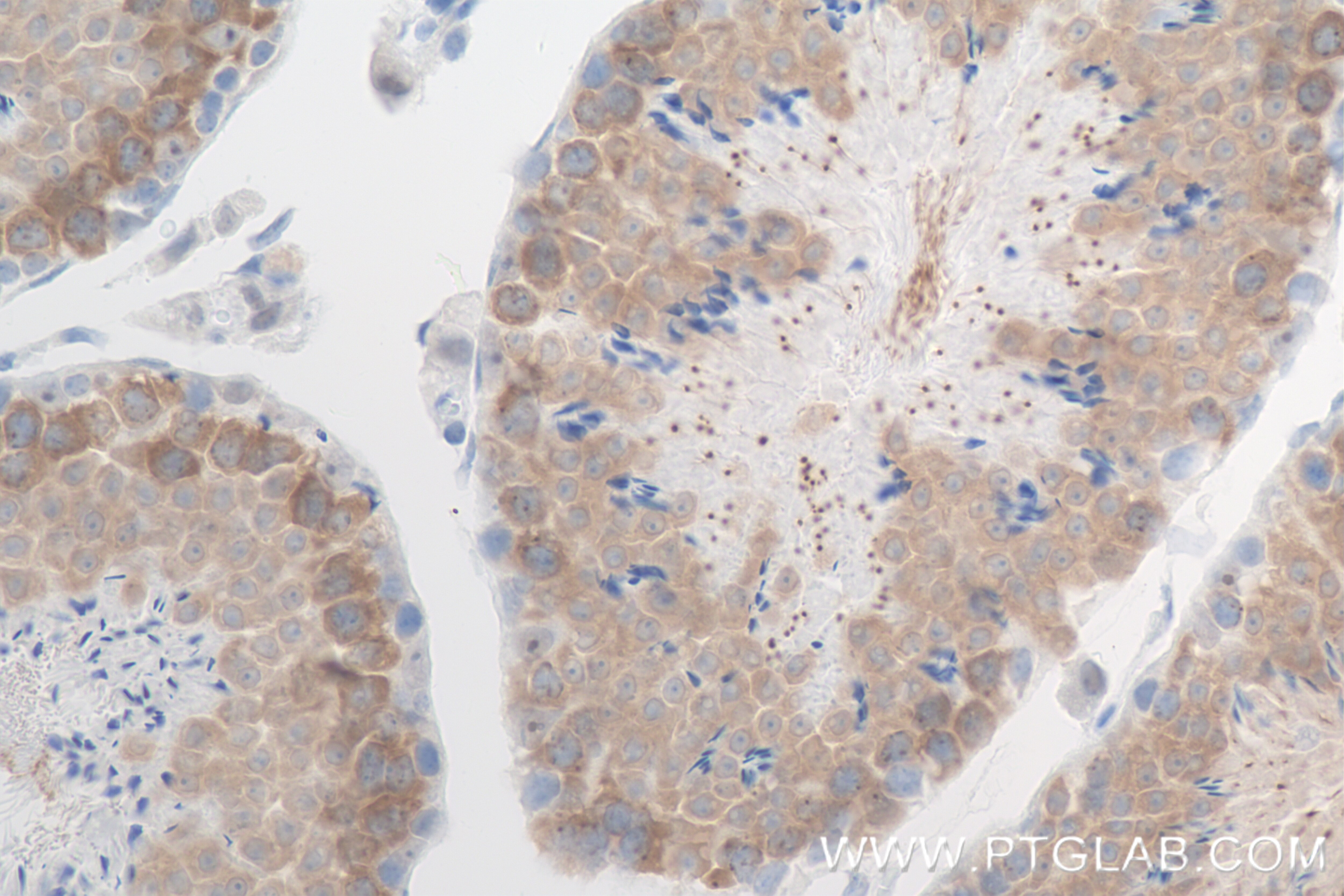
Tested Applications
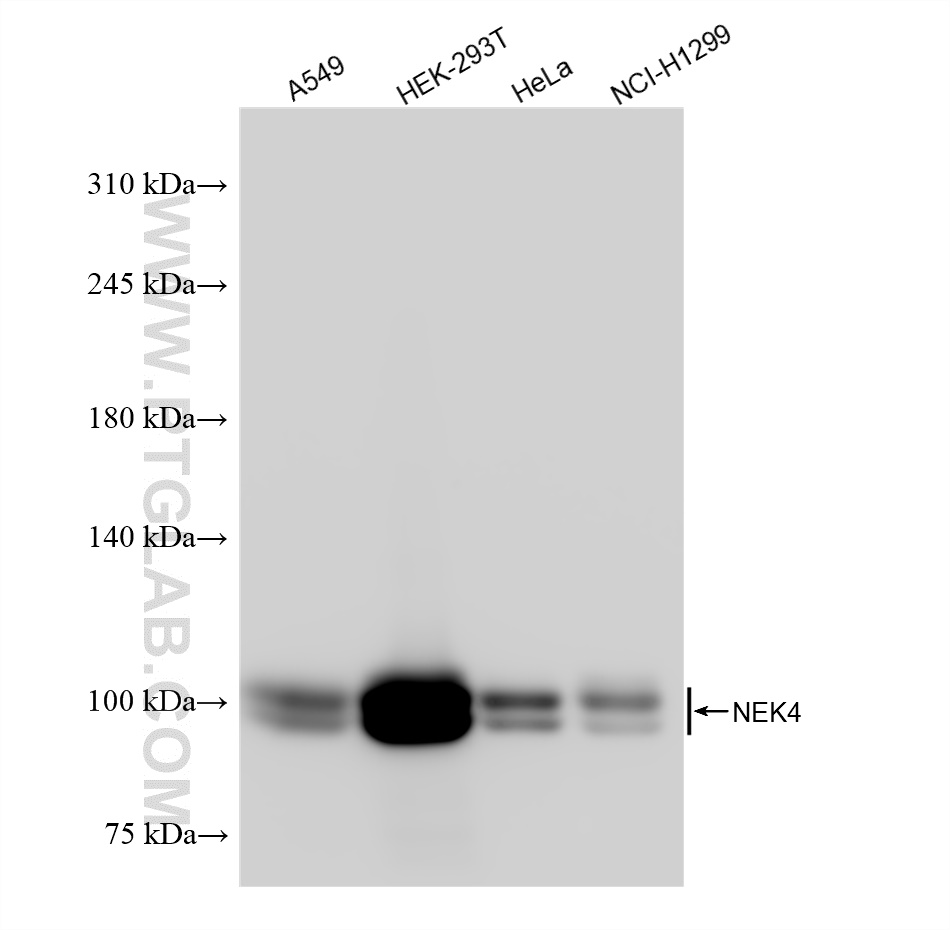
| Positive WB detected in | A549 cells, HEK-293T cells, HeLa cells, NCI-H1299 cells |
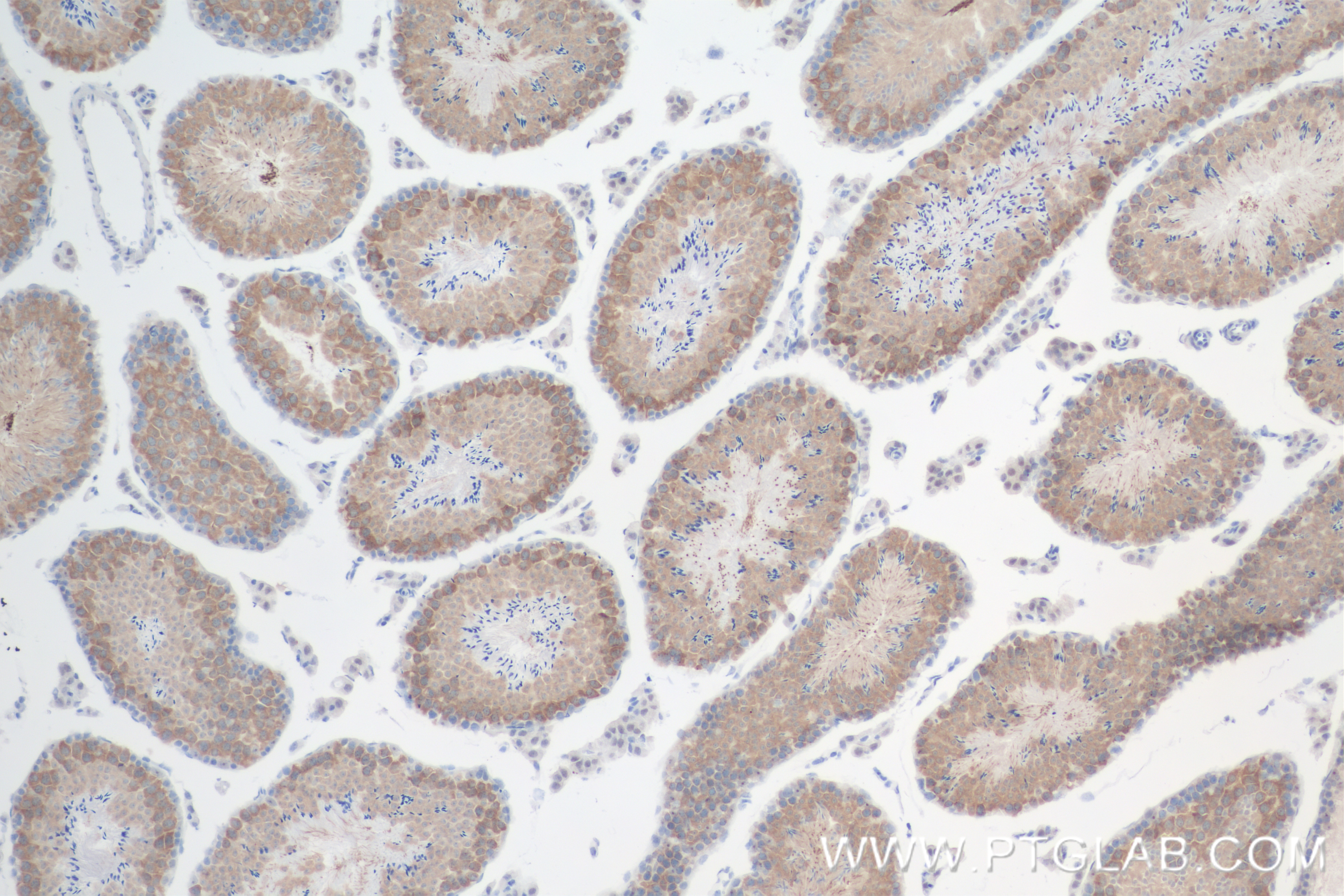
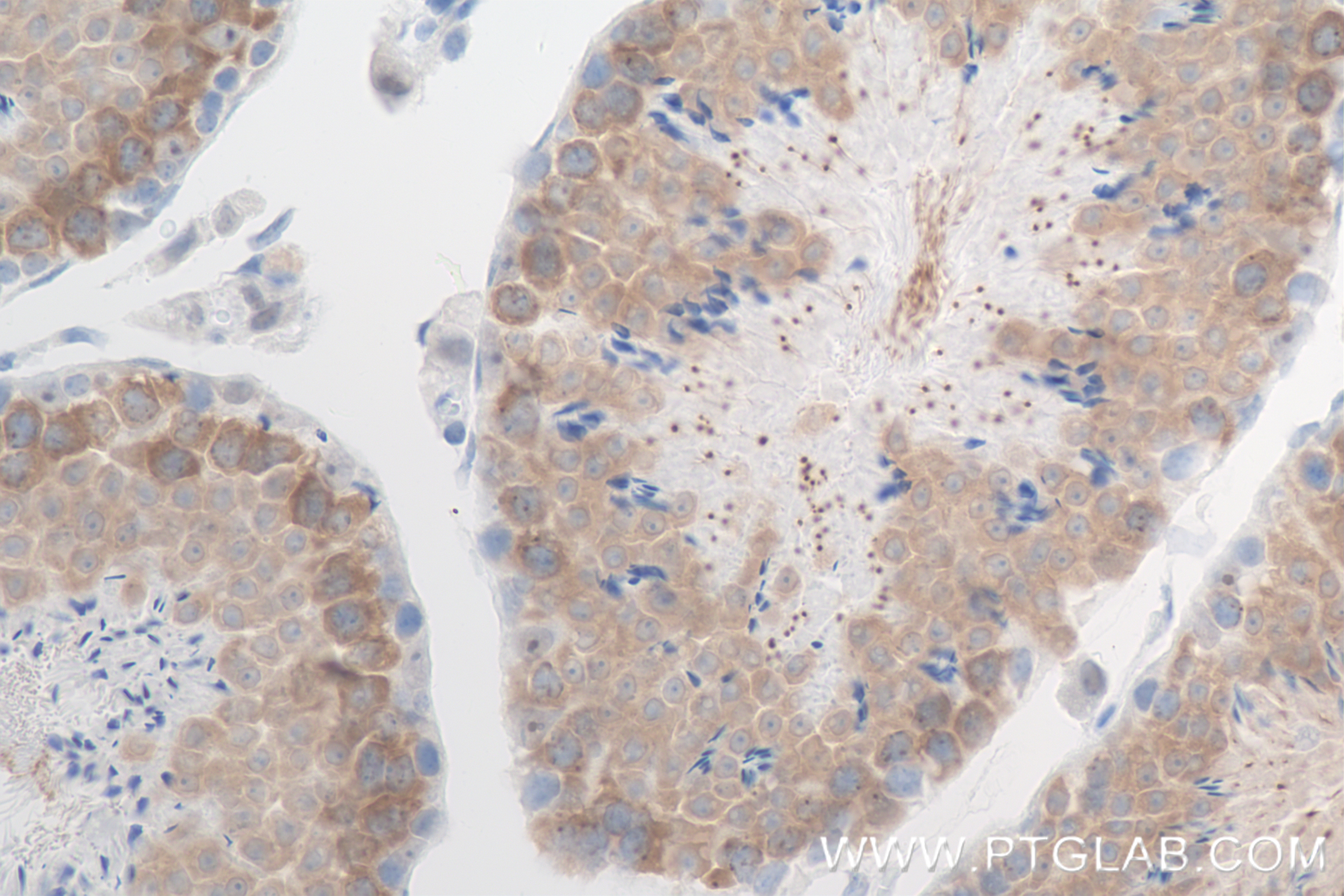
| Positive IHC detected in | mouse testis tissue Note: suggested antigen retrieval with TE buffer pH 9.0; (*) Alternatively, antigen retrieval may be performed with citrate buffer pH 6.0 |
Recommended dilution
| Application | Dilution |
|---|---|
| Western Blot (WB) | WB : 1:1000-1:8000 |
| Immunohistochemistry (IHC) | IHC : 1:250-1:1000 |
| It is recommended that this reagent should be titrated in each testing system to obtain optimal results. | |
| Sample-dependent, Check data in validation data gallery. | |
Product Information
84654-4-RR targets NEK4 in WB, IHC, ELISA applications and shows reactivity with human, mouse samples.
| Tested Reactivity | human, mouse |
| Host / Isotype | Rabbit / IgG |
| Class | Recombinant |
| Type | Antibody |
| Immunogen | NEK4 fusion protein Ag34929 Predict reactive species |
| Full Name | NIMA (never in mitosis gene a)-related kinase 4 |
| Calculated Molecular Weight | 781 aa, 88 kDa |
| Observed Molecular Weight | 95-100 kDa |
| GenBank Accession Number | BC063044 |
| Gene Symbol | NEK4 |
| Gene ID (NCBI) | 6787 |
| Conjugate | Unconjugated |
| Form | Liquid |
| Purification Method | Protein A purfication |
| UNIPROT ID | P51957 |
| Storage Buffer | PBS with 0.02% sodium azide and 50% glycerol , pH 7.3 |
| Storage Conditions | Store at -20°C. Stable for one year after shipment. Aliquoting is unnecessary for -20oC storage. 20ul sizes contain 0.1% BSA. |
Background Information
Neks (NIMA-related kinases) are a group of serine-threonine kinases that are related to NIMA, their ortholog from Aspergillus nidulans, which is essential for cells to enter in mitosis. NEK4 was initially identified as STK2, from serine/threonine kinase 2, in a study using a kinase specific cDNA library from human breast cancer tumors or breast cancer cells.
Protocols
| Product Specific Protocols | |
|---|---|
| WB protocol for NEK4 antibody 84654-4-RR | Download protocol |
| IHC protocol for NEK4 antibody 84654-4-RR | Download protocol |
| Standard Protocols | |
|---|---|
| Click here to view our Standard Protocols |