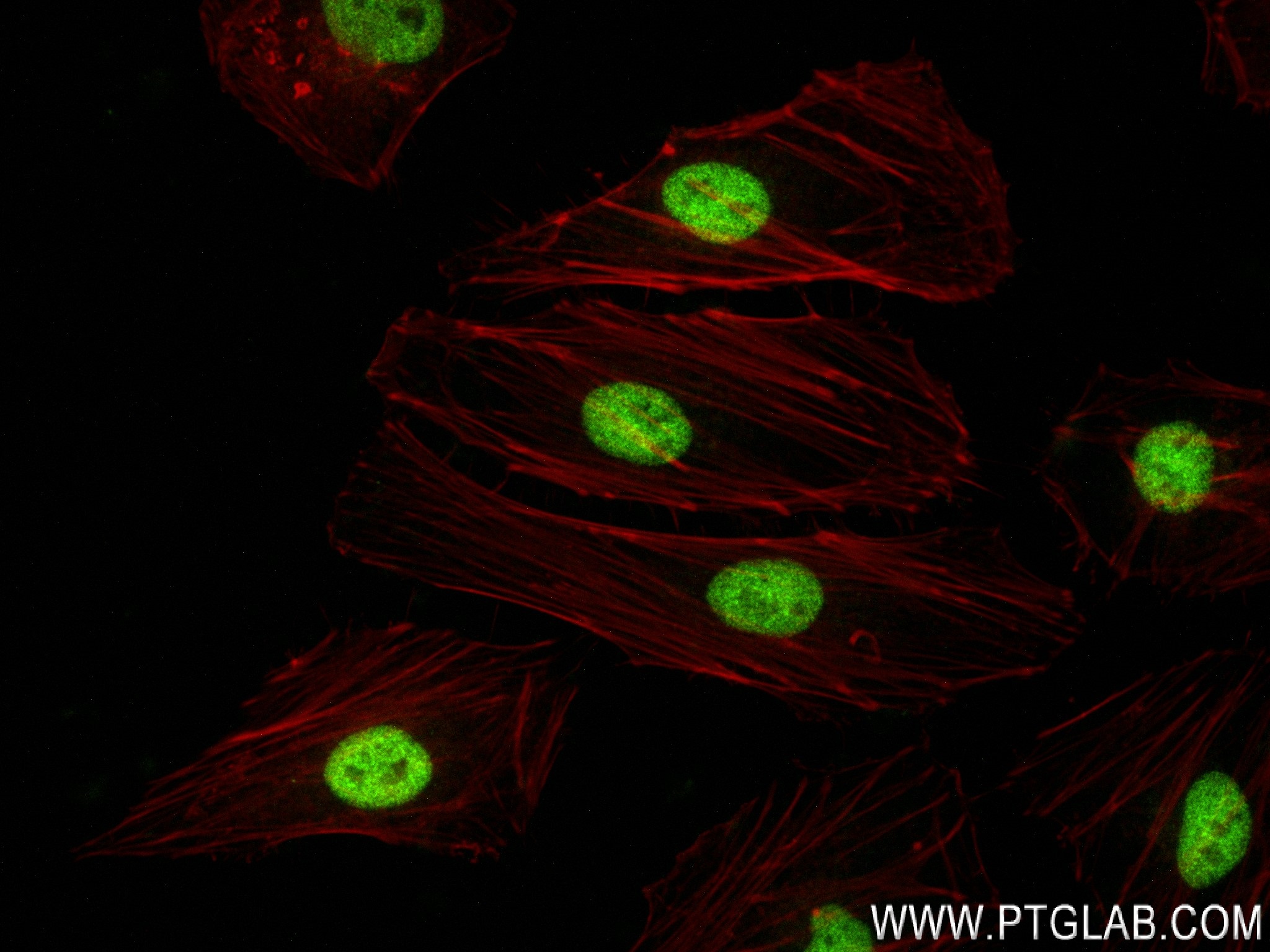
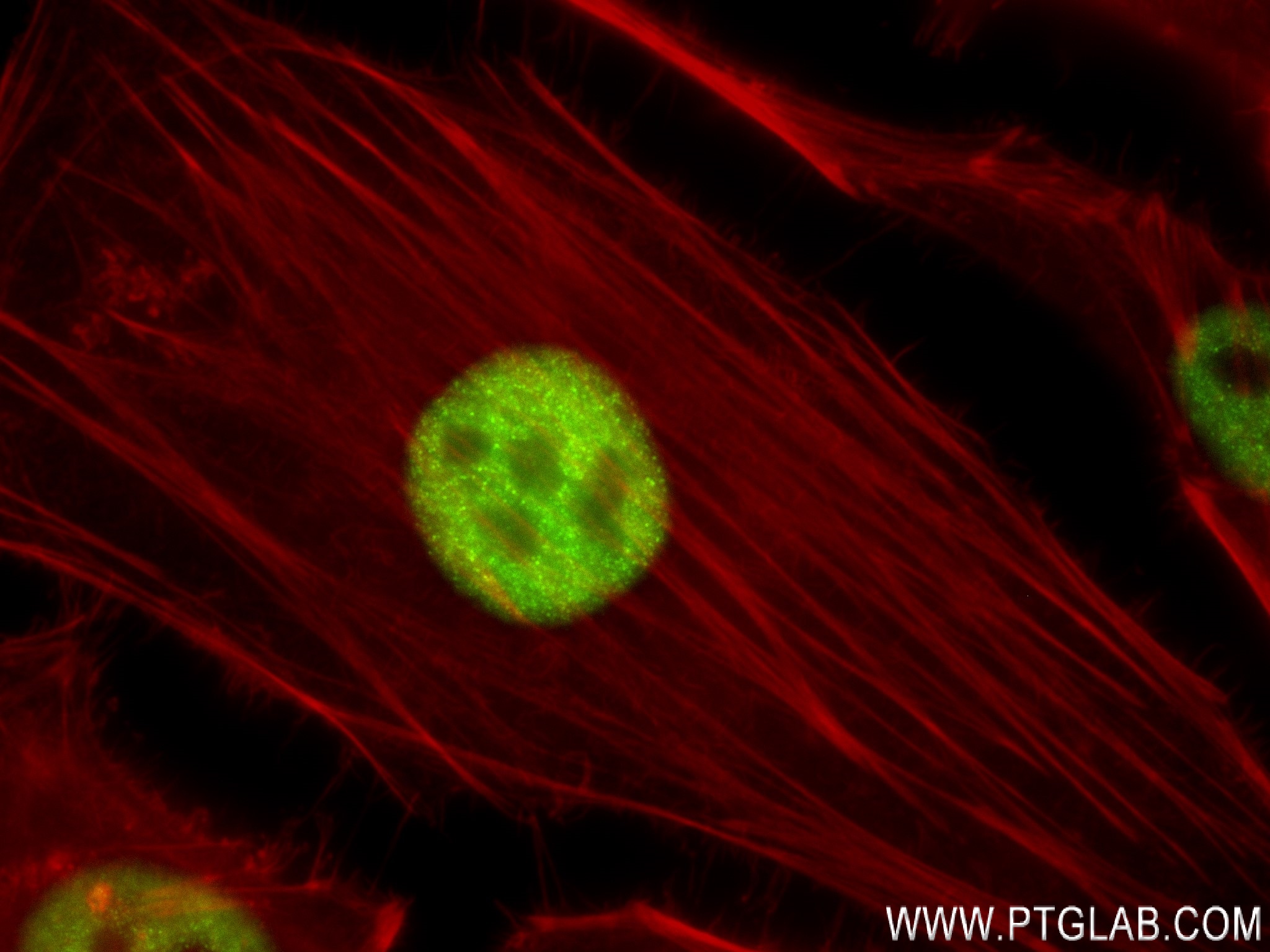
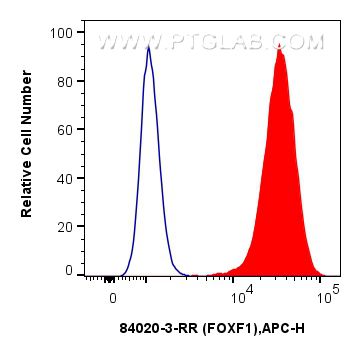
Tested Applications
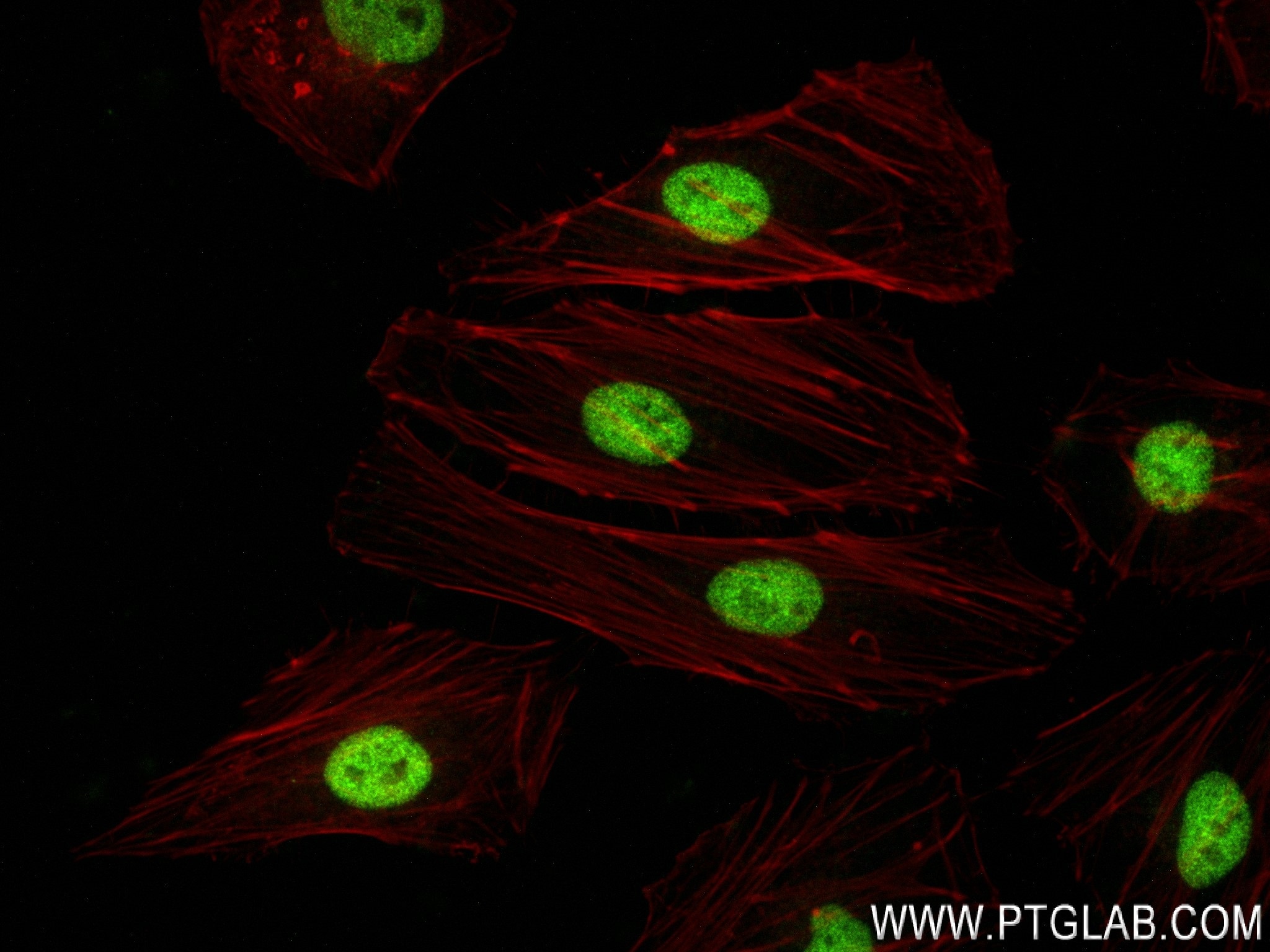
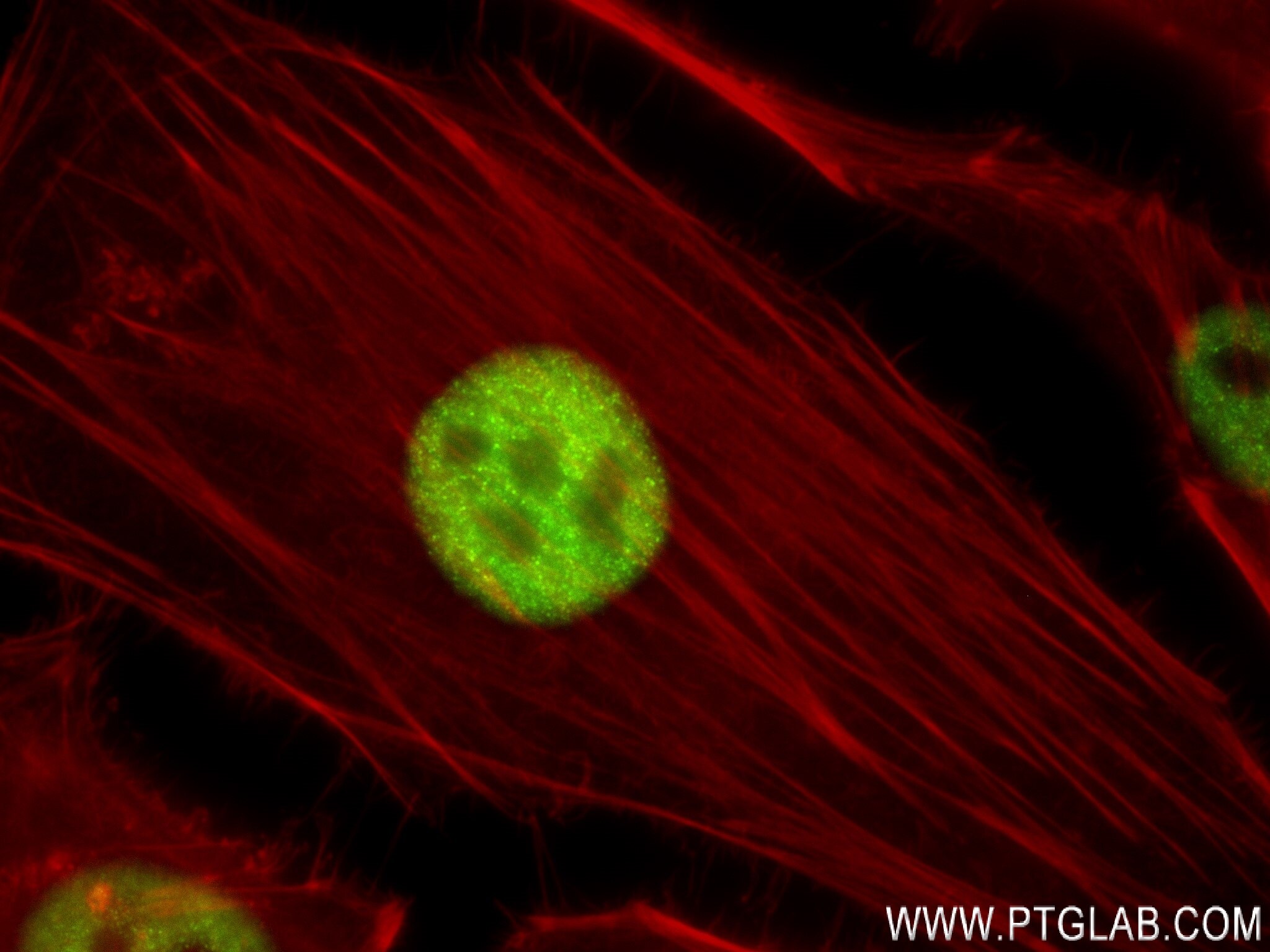
| Positive IF/ICC detected in | HeLa cells |
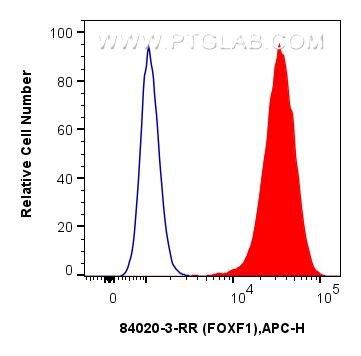
| Positive FC (Intra) detected in | HeLa cells |
Recommended dilution
| Application | Dilution |
|---|---|
| Immunofluorescence (IF)/ICC | IF/ICC : 1:125-1:500 |
| Flow Cytometry (FC) (INTRA) | FC (INTRA) : 0.25 ug per 10^6 cells in a 100 µl suspension |
| It is recommended that this reagent should be titrated in each testing system to obtain optimal results. | |
| Sample-dependent, Check data in validation data gallery. | |
Product Information
84020-3-RR targets FOXF1 in IF/ICC, FC (Intra), ELISA applications and shows reactivity with human samples.
| Tested Reactivity | human |
| Host / Isotype | Rabbit / IgG |
| Class | Recombinant |
| Type | Antibody |
| Immunogen | FOXF1 fusion protein Ag34266 Predict reactive species |
| Full Name | forkhead box F1 |
| Calculated Molecular Weight | 40 kDa |
| Observed Molecular Weight | 40 kDa |
| GenBank Accession Number | NP_001442 |
| Gene Symbol | FOXF1 |
| Gene ID (NCBI) | 2294 |
| RRID | AB_3671589 |
| Conjugate | Unconjugated |
| Form | Liquid |
| Purification Method | Protein A purfication |
| UNIPROT ID | Q12946 |
| Storage Buffer | PBS with 0.02% sodium azide and 50% glycerol , pH 7.3 |
| Storage Conditions | Store at -20°C. Stable for one year after shipment. Aliquoting is unnecessary for -20oC storage. 20ul sizes contain 0.1% BSA. |
Background Information
FOXF1 (Forkhead box protein F1), also known as FKHL5, It is expected to be located in the nucleus, which is expressed predominantly in lung and placenta. FOXF1, a member of the Forkhead box family of transcription factors, as one of the most highly and consistently upregulated genes in FP-RMS. While it is known that FOXF1 is required for mesodermal differentiation toward endothelial and smooth muscle cell lineages, FOXF1 is not expressed in differentiated skeletal muscle cells, and its role in fusion-positive FP-RMS remains unknown (PMID: 33627785). The molecular weight of FOXF1 is 40 kDa.
Protocols
| Product Specific Protocols | |
|---|---|
| IF protocol for FOXF1 antibody 84020-3-RR | Download protocol |
| Standard Protocols | |
|---|---|
| Click here to view our Standard Protocols |