Tested Applications
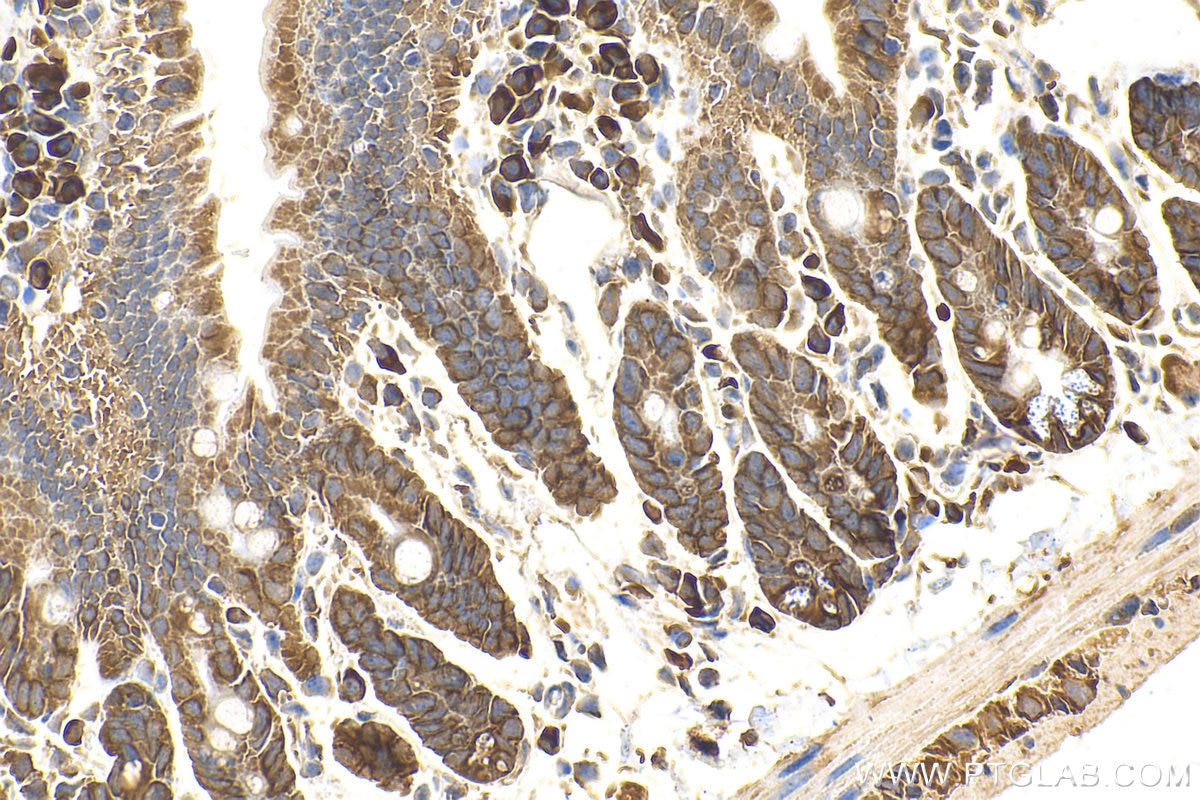
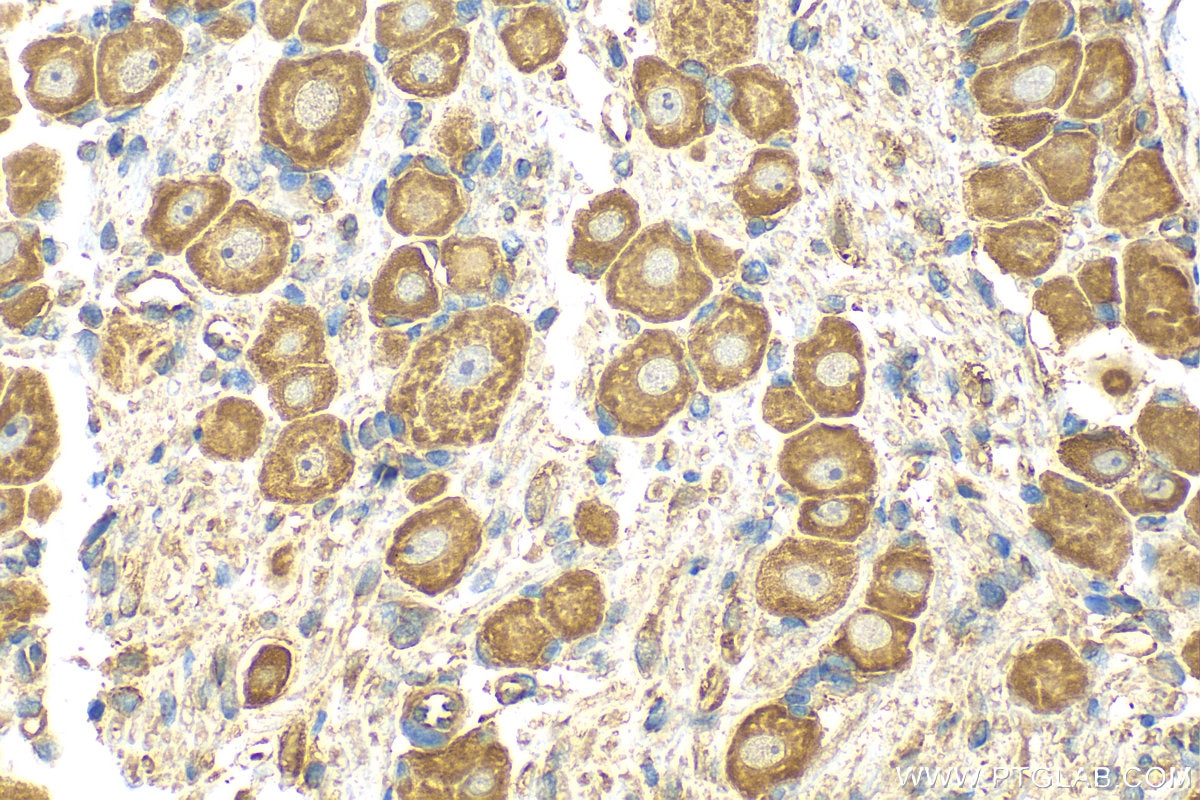
| Positive IHC detected in | mouse small intestine tissue, rat dorsal root ganglion tissue Note: suggested antigen retrieval with TE buffer pH 9.0; (*) Alternatively, antigen retrieval may be performed with citrate buffer pH 6.0 |
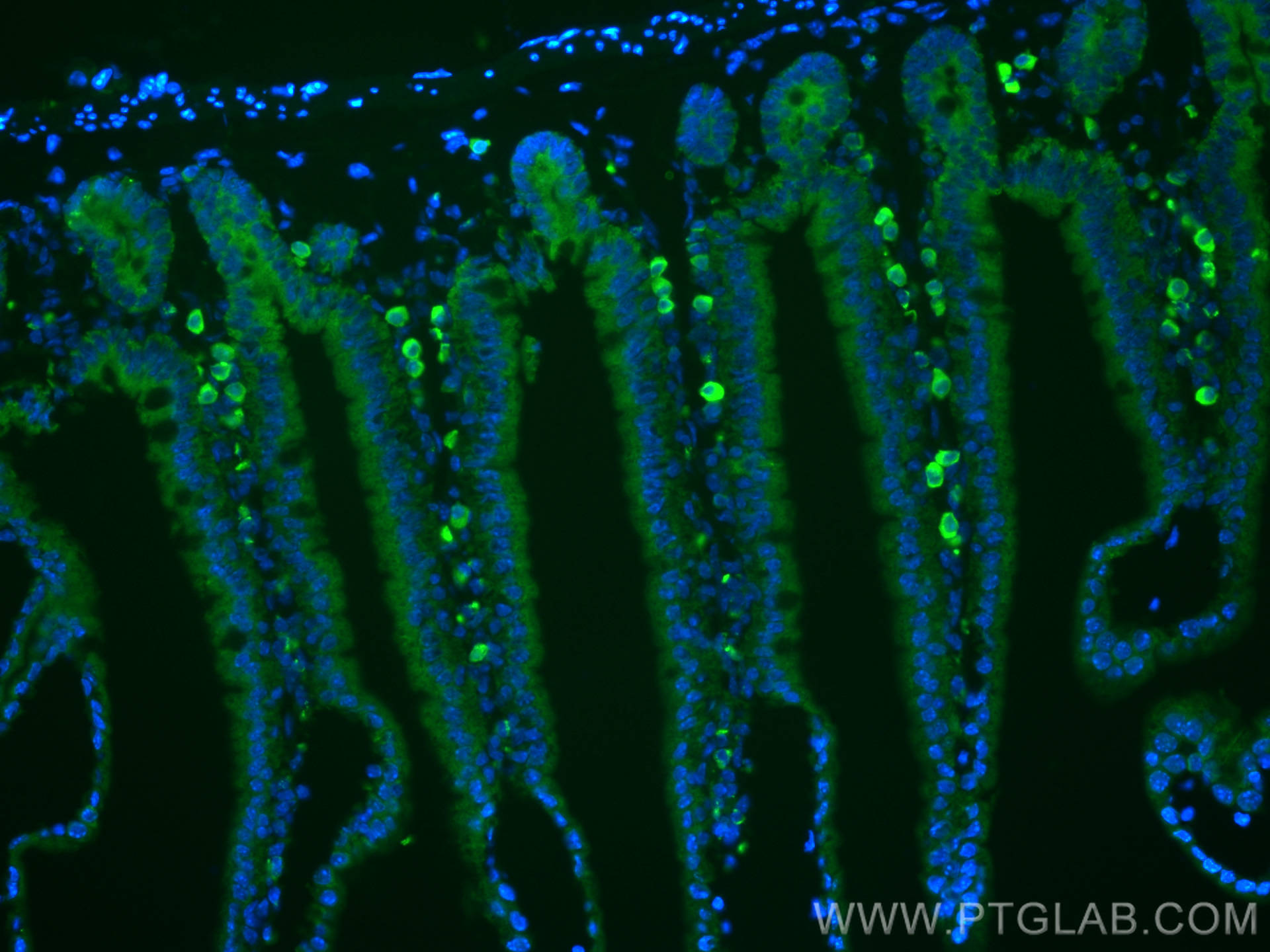
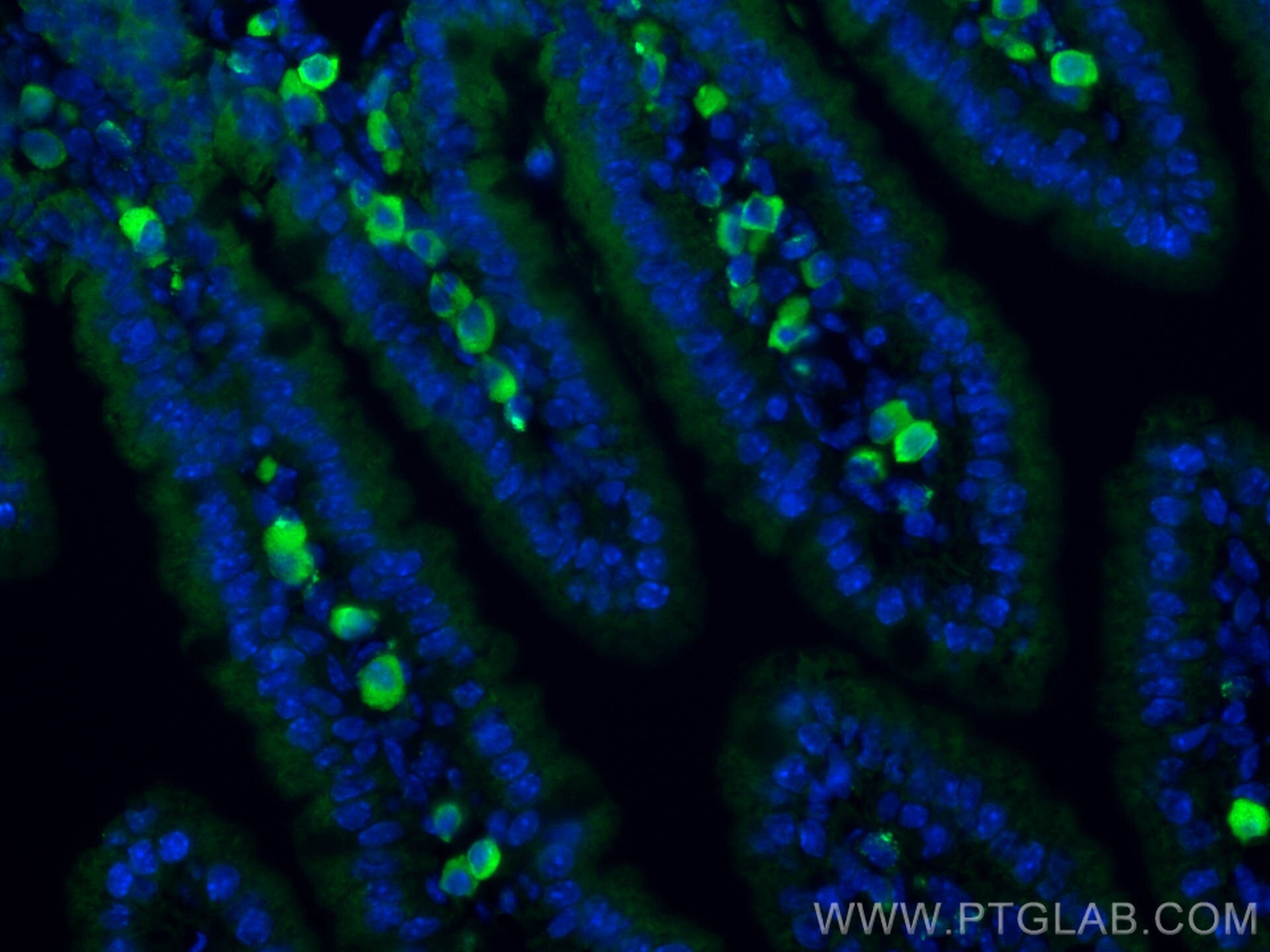
| Positive IF-P detected in | mouse small intestine tissue |
Recommended dilution
| Application | Dilution |
|---|---|
| Immunohistochemistry (IHC) | IHC : 1:50-1:500 |
| Immunofluorescence (IF)-P | IF-P : 1:200-1:800 |
| It is recommended that this reagent should be titrated in each testing system to obtain optimal results. | |
| Sample-dependent, Check data in validation data gallery. | |
Product Information
23640-1-AP targets MRGPRD in IHC, IF-P, ELISA applications and shows reactivity with human, mouse, rat samples.
| Tested Reactivity | human, mouse, rat |
| Host / Isotype | Rabbit / IgG |
| Class | Polyclonal |
| Type | Antibody |
| Immunogen | MRGPRD fusion protein Ag20316 Predict reactive species |
| Full Name | MAS-related GPR, member D |
| Calculated Molecular Weight | 321 aa, 36 kDa |
| GenBank Accession Number | BC156660 |
| Gene Symbol | MRGPRD |
| Gene ID (NCBI) | 116512 |
| RRID | AB_3669414 |
| Conjugate | Unconjugated |
| Form | Liquid |
| Purification Method | Antigen Affinity purified |
| UNIPROT ID | Q8TDS7 |
| Storage Buffer | PBS with 0.02% sodium azide and 50% glycerol pH 7.3. |
| Storage Conditions | Store at -20°C. Stable for one year after shipment. Aliquoting is unnecessary for -20oC storage. 20ul sizes contain 0.1% BSA. |
Background Information
MRGPRD (MAS-related G-protein coupled receptor, D), also referred to as hGPCR45 or TGR7, was identified as a novel GPCR in murine and human genomes. As for MRGD, its expression was found in dorsal root ganglia (DRG) and co-localized with Vanilloid receptor-1 (VR-1), which is an essential receptor for heat and pain sensation (PMID: 22715397).
Protocols
| Product Specific Protocols | |
|---|---|
| IHC protocol for MRGPRD antibody 23640-1-AP | Download protocol |
| IF protocol for MRGPRD antibody 23640-1-AP | Download protocol |
| Standard Protocols | |
|---|---|
| Click here to view our Standard Protocols |