Virus research using GFP
References from the last 4 years in virus research using GFP and ChromoTek's GFP-Trap
An extract of the published literature since 2016
This blog provides references from the last 4 years in virus research using GFP. Most publications report how immunoprecipitation (IP)/Co-IP of GFP-fusions was conducted to identify host cell binding partners of virus proteins. In addition, mass spectrometry analysis and functional assays have been performed.
In earlier research on corona viruses (not CoVID19, but previous viruses of the corona strain) our GFP-Trap has been applied. The results of the research work have been published in these references:
- p53 down-regulates SARS coronavirus replication and is targeted by the SARS-unique domain and PLpro via E3 ubiquitin ligase RCHY1. Y. Ma-Lauer, J. Carbajo-Lozoya, M. Y. Hein, M. A. Muller, W. Deng, J. Lei, B. Meyer, Y. Kusov, B. von Brunn, D. R. Bairad, S. Hunten, C. Drosten, H. Hermeking, H. Leonhardt, M. Mann, R. Hilgenfeld, A. von Brunn; Proceedings of the National Academy of Sciences 2016, 113 (35), E5192-E5201 (Pubmed ID 27519799)
- Tubulins interact with porcine and human S proteins of the genus Alphacoronavirus and support successful assembly and release of infectious viral particles. T. Rudiger, P. Mayrhofer, Y. Ma-Lauer, G. Pohlentz, J. Muthing, A. von Brunn, C. Schwegmann-Wessels; Virology 2016, 497, 185-1972016 (Pubmed ID 27479465)
- The cellular interactome of the coronavirus infectious bronchitis virus nucleocapsid protein and functional implications for virus biology. E. Emmott, D. Munday, E. Bickerton, P. Britton, M. A. Rodgers, A. Whitehouse, E. M. Zhou, J. A. Hiscox; Journal of Virology 2013, 87 (17), 9486-9500 (Pubmed ID 23637410)
This table shows an overview of literature references, in which the GFP-Trap has been used for virus research:
|
Virus Species |
Genus |
Family |
Application |
Pubmed ID / Link |
Year |
|
Abutilon mosaic virus (AbMV) |
Begomovirus |
Geminiviridae |
Co-IP, MS |
27693920 |
2017 |
|
Adenovirus |
n/a |
Adenoviridae |
Co-IP, MS |
27307198 |
2016 |
|
Alphacoronavirus |
Alphacoronavirus |
Coronaviridae |
Co-IP |
27479465 |
2016 |
|
Cauliflower mosaic virus |
Caulimovirus |
Caulimoviridae |
Co-IP |
30840696 |
2019 |
|
Chikungunya virus (CHIKV) |
Togaviridae |
Alphavirus |
Co-IP, MS |
29702546 |
2018 |
|
Chikungunya virus (CHIKV) |
Togaviridae |
Alphavirus |
IP, functional assays |
2020 |
|
|
Chikungunya virus (CHIKV) |
Togaviridae |
Alphavirus |
Co-IP |
31199850 |
2019 |
|
Citrus psorosis virus (CPsV) |
Ophiovirus |
Aspiviridae |
Co-IP |
30135122 |
2018 |
|
Cotton leaf curl Multan virus (CLCuMuV) |
Begomovirus |
Geminiviridae |
Co-IP |
27315204 |
2016 |
|
Cotton leaf curl Multan virus (CLCuMuV) |
Begomovirus |
Geminiviridae |
Co-IP |
28244873 |
2017 |
|
Dengue virus (DENV) |
Flavivirus |
Flaviviridae |
Co-IP |
27076639 |
2016 |
|
Dengue virus (DENV) |
Flavivirus |
Flaviviridae |
Co-IP, MS |
30608919 |
2019 |
|
Ebola virus |
Ebolavirus |
Filoviridae |
Co-IP |
29290611 |
2018 |
|
Ebola virus (EBOV) |
Ebolavirus |
Filoviridae |
Co-IP, MS |
27786485 |
2016 |
|
Enterovirus |
Enterovirus |
Picornaviridae |
Co-IP |
31381608 |
2019 |
|
Guinea pig cytomegalovirus (GPCMV) |
Cytomegalovirus |
Betaherpesvirinae |
Co-IP |
28651121 |
2017 |
|
Guinea pig cytomegalovirus (GPCMV) |
Cytomegalovirus |
Betaherpesvirinae |
Co-IP |
27387220 |
2016 |
|
Hendra virus V (HeV) |
Henipavirus |
Paramyxoviridae |
Co-IP |
29321677 |
2018 |
|
Hepatitis C virus (HCV) |
Hepacivirus |
Flaviviridae |
IP |
29352312 |
2018 |
|
Herpes simplex virus (HSV) |
Simplexvirus |
Herpesviridae |
Co-IP |
28507315 |
2017 |
|
Herpes simplex virus type 1 (HSV-1) |
Simplexvirus |
Herpesviridae |
Co-IP |
27852850 |
2017 |
|
Herpes simplex virus type 1 (HSV-1) |
Simplexvirus |
Herpesviridae |
Co-IP |
2019 |
|
|
Herpes simplex virus type 1 (HSV-1) |
Simplexvirus |
Herpesviridae |
Co-IP, MS |
2019 |
|
|
Herpes simplex virus type 1 (HSV-1) |
Simplexvirus |
Herpesviridae |
Co-IP, RIP |
28636603 |
2017 |
|
Herpes simplex virus type 2 (HSV-2) |
Simplexvirus |
Herpesviridae |
Co-IP |
30080903 |
2018 |
|
Human cytomegalovirus (HCMV) |
Cytomegalovirus |
Betaherpesvirinae |
Co-IP |
27181750 |
2016 |
|
Human immunodeficiency virus (HIV) |
Lentivirus |
Orthoretrovirinae |
Co-IP |
26792716 |
2016 |
|
Human immunodeficiency virus 1 (HIV-1) |
Lentivirus |
Orthoretrovirinae |
Co-IP |
28835492 |
2017 |
|
Human immunodeficiency virus 1 (HIV-1) |
Lentivirus |
Orthoretrovirinae |
Co-IP |
27560372 |
2016 |
|
Human immunodeficiency virus 1 (HIV-1) |
Lentivirus |
Orthoretrovirinae |
Co-IP |
31776272 |
2020 |
|
Human noroviruses (HuNoV) |
Norovirus |
Caliciviridae |
Co-IP |
31403400 |
2019 |
|
Human papillomavirus (HPV) |
n/a |
Papillomaviridae |
IP |
29045464 |
2017 |
|
Infectious bronchitis virus (IBV) |
Gammacoronavirus |
Coronaviridae |
Co-IP, MS |
23637410 |
2013 |
|
Influenza A virus |
Alphainfluenzavirus |
Orthomyxoviridae |
Co-IP |
29061883 |
2017 |
|
Influenza A virus |
Alphainfluenzavirus |
Orthomyxoviridae |
Co-IP |
28250123 |
2017 |
|
Kallithea virus (KV) |
Alphanudivirus |
Nudiviridae |
Co-IP, MS |
30404807 |
2019 |
|
Kaposi's sarcoma-associated herpesvirus (KSHV) |
Rhadinovirus |
Herpesviridae |
Co-IP |
27798559 |
2016 |
|
Kaposi's sarcoma-associated herpesvirus (KSHV) |
Rhadinovirus |
Herpesviridae |
Co-IP, MS |
26811480 |
2016 |
|
Merkel cell polyomavirus (MCPyV) |
Alphapolyomavirus |
Polyomaviridae |
Co-IP |
28445980 |
2017 |
|
Nipah virus (NiV) |
Henipavirus |
Paramyxoviridae |
Co-IP |
27622322 |
2016 |
|
Plum Pox Virus |
Potyvirus |
Potyviridae |
Co-IP |
27301551 |
2017 |
|
Porcine reproductive and respiratory syndrome virus (PRRSV) |
Porartevirus |
Arteriviridae |
Co-IP |
30819256 |
2019 |
|
Porcine reproductive and respiratory syndrome virus (PRRSV) |
Porartevirus |
Arteriviridae |
IP |
27327135 |
2016 |
|
Pseudorabies Virus |
Varicellovirus |
Herpesviridae |
ChIP |
28283558 |
2017 |
|
Rabies virus (RABV) |
Lyssavirus |
Rhabdoviridae |
Co-IP, AP-MS |
29084252 |
2017 |
|
Rabies virus (RABV) |
Lyssavirus |
Rhabdoviridae |
Co-IP |
26939125 |
2016 |
|
Red clover necrotic mosaic virus (RCNMV) |
Dianthovirus |
Tombusviridae |
Co-IP |
28154139 |
2017 |
|
Reptilian orthoreovirus |
Orthoreovirus |
Reoviridae |
Co-IP |
2019 |
|
|
Severe acute respiratory syndrome coronavirus (SARS-CoV) |
Betacoronavirus |
Coronaviridae |
Co-IP, ubiquitination/deubiquitination assay, kinase assay |
27519799 |
2016 |
|
Tick-borne encephalitis virus |
Flavivirus |
Flaviviridae |
Co-IP |
31243132 |
2019 |
|
Tobacco vein banding mosaic virus (TVBMV) |
Potyvirus |
Potyviridae |
Co-IP |
28230184 |
2017 |
|
Tomato yellow leaf curl virus (TYLCV) |
Begomovirus |
Geminiviridae |
Co-IP |
28946693 |
2017 |
|
Tomato yellow leaf curl virus (TYLCV) |
Begomovirus |
Geminiviridae |
Co-IP |
2020 |
|
|
Turnip mosaic virus (TuMV) |
Potyvirus |
Potyviridae |
Co-IP |
29593293 |
2018 |
|
Turnip mosaic virus (TuMV) |
Potyvirus |
Potyviridae |
Co-IP |
28223439 |
2017 |
|
Vaccinia virus (VacV) |
Orthopoxvirus |
Poxviridae |
Co-IP |
28467896 |
2017 |
|
Varicella zoster virus |
Varicellovirus |
Herpesviridae |
Co-IP |
2020 |
There are more references from 2015 and older, which are not included in above table. You can find those and additional publications, in which GFP-Trap has been applied to generate data, here.
ChromoTek’s GFP-Trap® is a versatile tool for the immunoprecipitation (IP) of GFP-tagged proteins. It consists of an anti-GFP Nanobody that is covalently coupled to beads. For a product overview see this link.
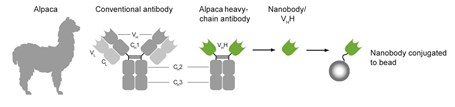
The interaction between the Nanobody and GFP is extraordinarily stable and efficient. This allows using cell lysate originating from many different organisms that require a multitude of different IP conditions and protocols. Due to the GFP-Trap’s high affinity and low background, low abundance GFP-fusion proteins can be effectively pulled down too.
Pictures: Many thanks for the virus picture to CDC on Unsplash
