Split fluorescent protein technology
The ChromoTek GFP-Trap has been successfully applied to multiple assays using different split GFP variants
ChromoTek scientists Michael Metterlein and Christian Linke-Winnebeck have published a whitepaper that provides a comprehensive overview with key developments in the field of split fluorescent protein technology. It also includes a selection of case studies on how ChromoTek’s Nano-Traps have been applied to exploit the full potential of this technology for example in protein-protein interaction studies. Particularly, the ChromoTek GFP-Trap has been successfully applied to multiple assays using different split GFP variants. Assay types include protein self-complementation, bimolecular fluorescence complementation (BiFC), tripartite fluorescence complementation (TriFC), and bimolecular complementation affinity purification (BiCAP).
The table below provides an overview of commonly used split fluorescent proteins (FPs); numbers after each split FP variant refer to respective β-strands included in fragments. Their split sites are mapped by colored stars to a topology diagram of GFP in the figure below.
A diagram of the secondary structure topology of GFP and its variants is shown below:
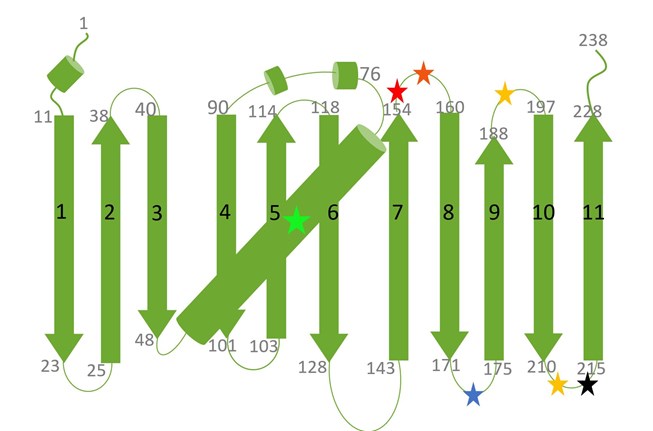
Presented are the 11 β-strands of GFP (or its variants) and the central α-helix. Black numbers on β-strands indicate the number of the respective β-strand, whereas the grey numbers on top or at the bottom of a β-strand mark individual amino acids (aa). The chromophore of any GFP variant is shown as a green star in the middle of the α-helix. Additional stars in red, blue, black and yellow mark split sites between β-strands for different split FP variants (see Table above for color codes).

